Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
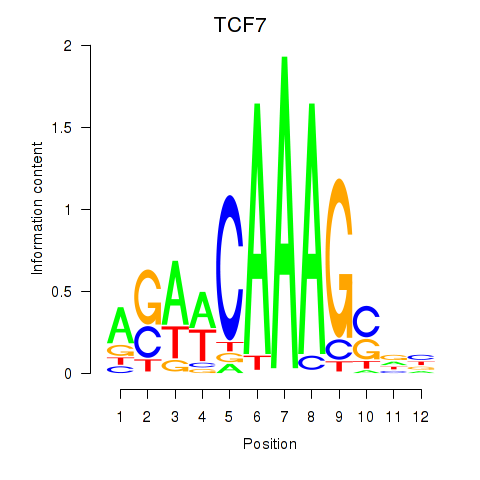
Results for TCF7
Z-value: 0.41
Transcription factors associated with TCF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7
|
ENSG00000081059.15 | transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7 | hg19_v2_chr5_+_133477905_133477956 | -0.47 | 3.4e-01 | Click! |
Activity profile of TCF7 motif
Sorted Z-values of TCF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_69080590 | 0.28 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr12_-_14924056 | 0.20 |
ENST00000539745.1
|
HIST4H4
|
histone cluster 4, H4 |
| chr2_-_99871570 | 0.15 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr1_+_59762642 | 0.14 |
ENST00000371218.4
ENST00000303721.7 |
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr7_+_95115210 | 0.14 |
ENST00000428113.1
ENST00000325885.5 |
ASB4
|
ankyrin repeat and SOCS box containing 4 |
| chr8_+_55471630 | 0.14 |
ENST00000522001.1
|
RP11-53M11.3
|
RP11-53M11.3 |
| chrX_-_148676974 | 0.13 |
ENST00000524178.1
|
HSFX2
|
heat shock transcription factor family, X linked 2 |
| chr19_-_44008863 | 0.12 |
ENST00000601646.1
|
PHLDB3
|
pleckstrin homology-like domain, family B, member 3 |
| chr11_+_12302492 | 0.12 |
ENST00000533534.1
|
MICALCL
|
MICAL C-terminal like |
| chrX_+_148855726 | 0.12 |
ENST00000370416.4
|
HSFX1
|
heat shock transcription factor family, X linked 1 |
| chr3_+_118892411 | 0.12 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr7_+_104581389 | 0.12 |
ENST00000415513.1
ENST00000417026.1 |
RP11-325F22.2
|
RP11-325F22.2 |
| chr10_-_104211294 | 0.11 |
ENST00000239125.1
|
C10orf95
|
chromosome 10 open reading frame 95 |
| chr19_+_12273866 | 0.10 |
ENST00000425827.1
ENST00000439995.1 ENST00000343979.4 ENST00000398616.2 ENST00000418338.1 |
ZNF136
|
zinc finger protein 136 |
| chr6_+_35704804 | 0.10 |
ENST00000373869.3
|
ARMC12
|
armadillo repeat containing 12 |
| chr6_+_111408698 | 0.10 |
ENST00000368851.5
|
SLC16A10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr6_-_30684898 | 0.09 |
ENST00000422266.1
ENST00000416571.1 |
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr10_+_7745232 | 0.09 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr11_-_3013482 | 0.09 |
ENST00000529361.1
ENST00000528968.1 ENST00000534372.1 ENST00000531291.1 ENST00000526842.1 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr4_-_140477910 | 0.09 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr12_-_58329819 | 0.09 |
ENST00000551421.1
|
RP11-620J15.3
|
RP11-620J15.3 |
| chr4_-_87515202 | 0.09 |
ENST00000502302.1
ENST00000513186.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr11_-_85430163 | 0.09 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr1_-_219615984 | 0.09 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr10_-_14614311 | 0.09 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr1_+_145470504 | 0.08 |
ENST00000323397.4
|
ANKRD34A
|
ankyrin repeat domain 34A |
| chr12_-_71003568 | 0.08 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr8_-_134511587 | 0.08 |
ENST00000523855.1
ENST00000523854.1 |
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr15_+_63414760 | 0.08 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr22_+_39052632 | 0.08 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr11_-_125366018 | 0.08 |
ENST00000527534.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr3_-_125775629 | 0.07 |
ENST00000383598.2
|
SLC41A3
|
solute carrier family 41, member 3 |
| chrX_-_55020511 | 0.07 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr4_-_74486347 | 0.07 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr10_-_14614095 | 0.07 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr7_+_140774032 | 0.07 |
ENST00000565468.1
|
TMEM178B
|
transmembrane protein 178B |
| chr17_+_48503519 | 0.07 |
ENST00000300441.4
ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr6_+_116937636 | 0.07 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr6_-_15548591 | 0.07 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr7_-_30009542 | 0.07 |
ENST00000438497.1
|
SCRN1
|
secernin 1 |
| chr5_-_36301984 | 0.07 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr10_-_99393208 | 0.07 |
ENST00000307450.6
|
MORN4
|
MORN repeat containing 4 |
| chr13_+_111855399 | 0.07 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr15_-_44487408 | 0.07 |
ENST00000402883.1
ENST00000417257.1 |
FRMD5
|
FERM domain containing 5 |
| chr12_+_104359614 | 0.07 |
ENST00000266775.9
ENST00000544861.1 |
TDG
|
thymine-DNA glycosylase |
| chr15_+_64752927 | 0.07 |
ENST00000416172.1
|
ZNF609
|
zinc finger protein 609 |
| chr4_-_100484825 | 0.07 |
ENST00000273962.3
ENST00000514547.1 ENST00000455368.2 |
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr1_+_244998918 | 0.07 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr14_+_32798462 | 0.07 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr10_-_14614122 | 0.06 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chr3_-_197676740 | 0.06 |
ENST00000452735.1
ENST00000453254.1 ENST00000455191.1 |
IQCG
|
IQ motif containing G |
| chr19_-_48389651 | 0.06 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr10_-_14613968 | 0.06 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr17_+_34848049 | 0.06 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr6_+_43968306 | 0.06 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr4_-_85419603 | 0.06 |
ENST00000295886.4
|
NKX6-1
|
NK6 homeobox 1 |
| chr19_+_57791419 | 0.06 |
ENST00000537645.1
|
ZNF460
|
zinc finger protein 460 |
| chr15_-_90294523 | 0.06 |
ENST00000300057.4
|
MESP1
|
mesoderm posterior 1 homolog (mouse) |
| chrX_+_100645812 | 0.05 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr5_-_157002775 | 0.05 |
ENST00000257527.4
|
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr16_+_2198604 | 0.05 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr11_-_3013316 | 0.05 |
ENST00000430811.1
|
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr10_+_96698406 | 0.05 |
ENST00000260682.6
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr7_-_102232891 | 0.05 |
ENST00000514917.2
|
RP11-514P8.7
|
RP11-514P8.7 |
| chr12_+_32654965 | 0.05 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr15_-_52043722 | 0.05 |
ENST00000454181.2
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr9_-_35665165 | 0.05 |
ENST00000343259.3
ENST00000378387.3 |
ARHGEF39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr4_+_159593271 | 0.05 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr20_+_35504522 | 0.05 |
ENST00000602922.1
ENST00000217320.3 |
TLDC2
|
TBC/LysM-associated domain containing 2 |
| chr17_-_74489215 | 0.05 |
ENST00000585701.1
ENST00000591192.1 ENST00000589526.1 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr10_+_7745303 | 0.05 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr11_-_111383064 | 0.05 |
ENST00000525791.1
ENST00000456861.2 ENST00000356018.2 |
BTG4
|
B-cell translocation gene 4 |
| chr6_+_88054530 | 0.05 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chrX_-_15332665 | 0.05 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr18_+_12407895 | 0.05 |
ENST00000590956.1
ENST00000336990.4 ENST00000440960.1 ENST00000588729.1 |
SLMO1
|
slowmo homolog 1 (Drosophila) |
| chr7_+_23286182 | 0.05 |
ENST00000258733.4
ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB
|
glycoprotein (transmembrane) nmb |
| chr9_+_103189405 | 0.05 |
ENST00000395067.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr2_-_217560248 | 0.05 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr16_+_8806800 | 0.05 |
ENST00000561870.1
ENST00000396600.2 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr10_+_54074033 | 0.05 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr1_+_110009215 | 0.05 |
ENST00000369872.3
|
SYPL2
|
synaptophysin-like 2 |
| chr8_+_97773457 | 0.05 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr7_+_123469037 | 0.05 |
ENST00000489978.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr10_+_70847852 | 0.04 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr2_-_231084659 | 0.04 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chrX_+_120181457 | 0.04 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr3_+_36421826 | 0.04 |
ENST00000273183.3
|
STAC
|
SH3 and cysteine rich domain |
| chr22_+_19950060 | 0.04 |
ENST00000449653.1
|
COMT
|
catechol-O-methyltransferase |
| chr11_-_85430088 | 0.04 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr4_-_7069760 | 0.04 |
ENST00000264954.4
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli) |
| chr22_+_39077947 | 0.04 |
ENST00000216034.4
|
TOMM22
|
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr16_-_46864955 | 0.04 |
ENST00000565112.1
|
C16orf87
|
chromosome 16 open reading frame 87 |
| chr18_+_29769978 | 0.04 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chrX_+_100646190 | 0.04 |
ENST00000471855.1
|
RPL36A
|
ribosomal protein L36a |
| chr12_+_4714145 | 0.04 |
ENST00000545342.1
|
DYRK4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr8_+_56792355 | 0.04 |
ENST00000519728.1
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr6_-_25874440 | 0.04 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr19_+_41256764 | 0.04 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr3_-_167813672 | 0.04 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr7_-_148725733 | 0.04 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr12_-_49581152 | 0.04 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr4_+_95972822 | 0.04 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr7_-_135433460 | 0.04 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr2_-_27592867 | 0.04 |
ENST00000451130.2
|
EIF2B4
|
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr22_+_30821732 | 0.04 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr17_-_18266818 | 0.04 |
ENST00000583780.1
|
SHMT1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr3_+_197677379 | 0.04 |
ENST00000442341.1
|
RPL35A
|
ribosomal protein L35a |
| chr8_+_30244580 | 0.04 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr17_-_63557309 | 0.04 |
ENST00000580513.1
|
AXIN2
|
axin 2 |
| chr20_-_17511962 | 0.04 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr15_+_36871806 | 0.04 |
ENST00000566621.1
ENST00000564586.1 |
C15orf41
|
chromosome 15 open reading frame 41 |
| chr1_-_213020991 | 0.04 |
ENST00000332912.3
|
C1orf227
|
chromosome 1 open reading frame 227 |
| chr12_+_104359641 | 0.04 |
ENST00000537100.1
|
TDG
|
thymine-DNA glycosylase |
| chr17_+_41150290 | 0.04 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr12_-_109915098 | 0.04 |
ENST00000542858.1
ENST00000542262.1 ENST00000424763.2 |
KCTD10
|
potassium channel tetramerization domain containing 10 |
| chr3_+_57875711 | 0.04 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr5_+_34757309 | 0.04 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr17_-_34890037 | 0.04 |
ENST00000589404.1
|
MYO19
|
myosin XIX |
| chrX_-_138287168 | 0.04 |
ENST00000436198.1
|
FGF13
|
fibroblast growth factor 13 |
| chr7_-_35293740 | 0.04 |
ENST00000408931.3
|
TBX20
|
T-box 20 |
| chr7_-_16840820 | 0.04 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr21_+_30672433 | 0.04 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr14_+_61201445 | 0.04 |
ENST00000261245.4
ENST00000539616.2 |
MNAT1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr4_-_106629796 | 0.03 |
ENST00000416543.1
ENST00000515819.1 ENST00000420368.2 ENST00000503746.1 ENST00000340139.5 ENST00000433009.1 |
INTS12
|
integrator complex subunit 12 |
| chr7_-_27213893 | 0.03 |
ENST00000283921.4
|
HOXA10
|
homeobox A10 |
| chr8_+_56792377 | 0.03 |
ENST00000520220.2
|
LYN
|
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr3_+_111578131 | 0.03 |
ENST00000498699.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr10_+_75668916 | 0.03 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr7_+_134576317 | 0.03 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr3_+_111578027 | 0.03 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr19_+_13134772 | 0.03 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr7_+_150076406 | 0.03 |
ENST00000329630.5
|
ZNF775
|
zinc finger protein 775 |
| chr18_+_59992514 | 0.03 |
ENST00000269485.7
|
TNFRSF11A
|
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| chr5_+_125935960 | 0.03 |
ENST00000297540.4
|
PHAX
|
phosphorylated adaptor for RNA export |
| chrX_+_102883620 | 0.03 |
ENST00000372626.3
|
TCEAL1
|
transcription elongation factor A (SII)-like 1 |
| chr1_-_32801825 | 0.03 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr11_-_8615488 | 0.03 |
ENST00000315204.1
ENST00000396672.1 ENST00000396673.1 |
STK33
|
serine/threonine kinase 33 |
| chr11_+_100862811 | 0.03 |
ENST00000303130.2
|
TMEM133
|
transmembrane protein 133 |
| chr1_-_33502528 | 0.03 |
ENST00000354858.6
|
AK2
|
adenylate kinase 2 |
| chr4_-_146019693 | 0.03 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr18_+_3449821 | 0.03 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr4_+_56815102 | 0.03 |
ENST00000257287.4
|
CEP135
|
centrosomal protein 135kDa |
| chr3_+_118892362 | 0.03 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr12_-_49351148 | 0.03 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr5_-_174871136 | 0.03 |
ENST00000393752.2
|
DRD1
|
dopamine receptor D1 |
| chr12_+_78224667 | 0.03 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr17_+_30813576 | 0.03 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr15_-_48470544 | 0.03 |
ENST00000267836.6
|
MYEF2
|
myelin expression factor 2 |
| chr10_-_17171785 | 0.03 |
ENST00000377823.1
|
CUBN
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr3_+_52719936 | 0.03 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr3_-_169487617 | 0.03 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr2_-_74781061 | 0.03 |
ENST00000264094.3
ENST00000393937.2 ENST00000409986.1 |
LOXL3
|
lysyl oxidase-like 3 |
| chr1_-_155162658 | 0.03 |
ENST00000368389.2
ENST00000368396.4 ENST00000343256.5 ENST00000342482.4 ENST00000368398.3 ENST00000368390.3 ENST00000337604.5 ENST00000368392.3 ENST00000438413.1 ENST00000368393.3 ENST00000457295.2 ENST00000338684.5 ENST00000368395.1 |
MUC1
|
mucin 1, cell surface associated |
| chr8_-_42752418 | 0.03 |
ENST00000524954.1
|
RNF170
|
ring finger protein 170 |
| chr14_+_85996507 | 0.03 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_-_161695074 | 0.03 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr2_+_27593389 | 0.03 |
ENST00000233575.2
ENST00000543024.1 ENST00000537606.1 |
SNX17
|
sorting nexin 17 |
| chr6_-_112408661 | 0.03 |
ENST00000368662.5
|
TUBE1
|
tubulin, epsilon 1 |
| chrX_-_63425561 | 0.03 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr1_-_205649580 | 0.03 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr5_-_140700322 | 0.03 |
ENST00000313368.5
|
TAF7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr2_+_166326157 | 0.03 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr6_-_112575838 | 0.03 |
ENST00000455073.1
|
LAMA4
|
laminin, alpha 4 |
| chr2_+_159651821 | 0.03 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr16_+_30953696 | 0.03 |
ENST00000566320.2
ENST00000565939.1 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr8_-_41754231 | 0.03 |
ENST00000265709.8
|
ANK1
|
ankyrin 1, erythrocytic |
| chr5_+_32531893 | 0.03 |
ENST00000512913.1
|
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr17_-_46623441 | 0.03 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr4_-_7873713 | 0.03 |
ENST00000382543.3
|
AFAP1
|
actin filament associated protein 1 |
| chr2_-_27593180 | 0.03 |
ENST00000493344.2
ENST00000445933.2 |
EIF2B4
|
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr17_-_20946710 | 0.03 |
ENST00000584538.1
|
USP22
|
ubiquitin specific peptidase 22 |
| chr7_-_154863264 | 0.03 |
ENST00000395731.2
ENST00000543018.1 |
HTR5A-AS1
|
HTR5A antisense RNA 1 |
| chr2_-_86564696 | 0.03 |
ENST00000437769.1
|
REEP1
|
receptor accessory protein 1 |
| chr6_+_138188551 | 0.03 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr8_-_119964434 | 0.03 |
ENST00000297350.4
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b |
| chr1_-_151345159 | 0.03 |
ENST00000458566.1
ENST00000447402.3 ENST00000426705.2 ENST00000435071.1 ENST00000368868.5 |
SELENBP1
|
selenium binding protein 1 |
| chr1_-_33502441 | 0.03 |
ENST00000548033.1
ENST00000487289.1 ENST00000373449.2 ENST00000480134.1 ENST00000467905.1 |
AK2
|
adenylate kinase 2 |
| chr2_+_10183651 | 0.03 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr1_+_166808667 | 0.03 |
ENST00000537173.1
ENST00000536514.1 ENST00000449930.1 |
POGK
|
pogo transposable element with KRAB domain |
| chr2_-_128568721 | 0.03 |
ENST00000322313.4
ENST00000393006.1 ENST00000409658.3 ENST00000436787.1 |
WDR33
|
WD repeat domain 33 |
| chr1_+_20208870 | 0.03 |
ENST00000375120.3
|
OTUD3
|
OTU domain containing 3 |
| chr19_-_2328572 | 0.03 |
ENST00000252622.10
|
LSM7
|
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr11_+_45943169 | 0.03 |
ENST00000529052.1
ENST00000531526.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr1_-_11107280 | 0.03 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chr17_-_56065484 | 0.03 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr1_-_147142358 | 0.03 |
ENST00000392988.2
|
ACP6
|
acid phosphatase 6, lysophosphatidic |
| chr22_-_26908343 | 0.03 |
ENST00000407431.1
ENST00000455080.1 ENST00000418876.1 ENST00000440258.1 ENST00000407148.1 ENST00000420242.1 ENST00000405938.1 ENST00000407690.1 |
TFIP11
|
tuftelin interacting protein 11 |
| chr3_+_197677047 | 0.03 |
ENST00000448864.1
|
RPL35A
|
ribosomal protein L35a |
| chr11_-_6677018 | 0.02 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr1_+_183774285 | 0.02 |
ENST00000539189.1
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr21_+_33784670 | 0.02 |
ENST00000300255.2
|
EVA1C
|
eva-1 homolog C (C. elegans) |
| chr15_-_73076030 | 0.02 |
ENST00000311669.8
|
ADPGK
|
ADP-dependent glucokinase |
| chr4_+_2819883 | 0.02 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr3_-_25824353 | 0.02 |
ENST00000427041.1
|
NGLY1
|
N-glycanase 1 |
| chr1_+_156254070 | 0.02 |
ENST00000405535.2
ENST00000456810.1 |
TMEM79
|
transmembrane protein 79 |
| chr2_+_232573208 | 0.02 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr2_+_234580499 | 0.02 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr15_-_73075964 | 0.02 |
ENST00000563907.1
|
ADPGK
|
ADP-dependent glucokinase |
| chr15_+_74218787 | 0.02 |
ENST00000261921.7
|
LOXL1
|
lysyl oxidase-like 1 |
| chr4_-_186682716 | 0.02 |
ENST00000445343.1
|
SORBS2
|
sorbin and SH3 domain containing 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:0070666 | regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0021912 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) |
| 0.0 | 0.0 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.1 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.0 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.0 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.0 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.0 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.0 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |