Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
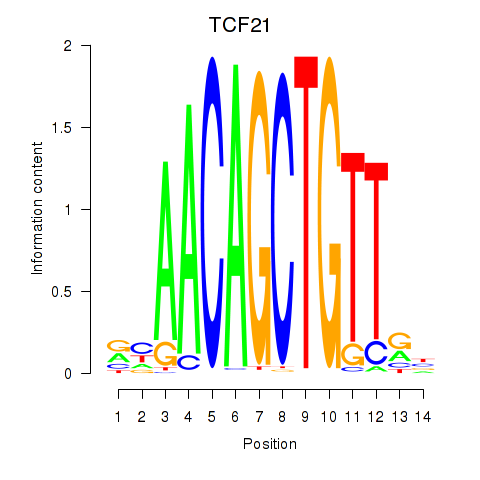
Results for TCF21
Z-value: 0.97
Transcription factors associated with TCF21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF21
|
ENSG00000118526.6 | transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF21 | hg19_v2_chr6_+_134210243_134210276 | 0.28 | 6.0e-01 | Click! |
Activity profile of TCF21 motif
Sorted Z-values of TCF21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_615570 | 2.09 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr1_+_79115503 | 1.51 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr11_-_615942 | 1.23 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr11_+_117103333 | 0.83 |
ENST00000534428.1
|
RNF214
|
ring finger protein 214 |
| chr2_+_231280954 | 0.79 |
ENST00000409824.1
ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100
|
SP100 nuclear antigen |
| chr2_+_231280908 | 0.71 |
ENST00000427101.2
ENST00000432979.1 |
SP100
|
SP100 nuclear antigen |
| chr1_+_92414952 | 0.56 |
ENST00000449584.1
ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT
|
bromodomain, testis-specific |
| chr3_+_167453493 | 0.53 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr6_-_41673552 | 0.48 |
ENST00000419574.1
ENST00000445214.1 |
TFEB
|
transcription factor EB |
| chr2_-_7005785 | 0.46 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr13_+_41635617 | 0.42 |
ENST00000542082.1
|
WBP4
|
WW domain binding protein 4 |
| chr1_+_172422026 | 0.41 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr17_+_71228793 | 0.40 |
ENST00000426147.2
|
C17orf80
|
chromosome 17 open reading frame 80 |
| chr17_+_71229346 | 0.40 |
ENST00000535032.2
ENST00000582793.1 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr8_-_99837856 | 0.40 |
ENST00000518165.1
ENST00000419617.2 |
STK3
|
serine/threonine kinase 3 |
| chr17_-_62499334 | 0.39 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr11_+_117103441 | 0.37 |
ENST00000531287.1
ENST00000531452.1 |
RNF214
|
ring finger protein 214 |
| chr10_+_52499682 | 0.37 |
ENST00000185907.9
ENST00000374006.1 |
ASAH2B
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2B |
| chr22_-_22863466 | 0.37 |
ENST00000406426.1
ENST00000360412.2 |
ZNF280B
|
zinc finger protein 280B |
| chr20_-_23402028 | 0.37 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chrM_+_7586 | 0.37 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr2_+_149402009 | 0.37 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr2_+_86116396 | 0.36 |
ENST00000455121.3
|
AC105053.4
|
AC105053.4 |
| chr8_-_71519889 | 0.36 |
ENST00000521425.1
|
TRAM1
|
translocation associated membrane protein 1 |
| chr19_-_33716750 | 0.34 |
ENST00000253188.4
|
SLC7A10
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10 |
| chr2_+_44001172 | 0.34 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr12_-_50677255 | 0.33 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr13_-_20767037 | 0.33 |
ENST00000382848.4
|
GJB2
|
gap junction protein, beta 2, 26kDa |
| chr10_-_105238997 | 0.32 |
ENST00000369783.4
|
CALHM3
|
calcium homeostasis modulator 3 |
| chr2_-_17981462 | 0.31 |
ENST00000402989.1
ENST00000428868.1 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr3_-_81792780 | 0.31 |
ENST00000489715.1
|
GBE1
|
glucan (1,4-alpha-), branching enzyme 1 |
| chr11_+_74862100 | 0.31 |
ENST00000532236.1
ENST00000531756.1 |
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr2_+_38177575 | 0.31 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr9_-_123812542 | 0.31 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr17_+_68165657 | 0.31 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr11_-_10920714 | 0.30 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr17_-_7193711 | 0.30 |
ENST00000571464.1
|
YBX2
|
Y box binding protein 2 |
| chr3_+_167453026 | 0.29 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr17_+_11924129 | 0.29 |
ENST00000353533.5
ENST00000415385.3 |
MAP2K4
|
mitogen-activated protein kinase kinase 4 |
| chr17_+_71228740 | 0.29 |
ENST00000268942.8
ENST00000359042.2 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr20_-_52790055 | 0.29 |
ENST00000395955.3
|
CYP24A1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr15_+_65337708 | 0.29 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr21_+_37692481 | 0.27 |
ENST00000400485.1
|
MORC3
|
MORC family CW-type zinc finger 3 |
| chr11_-_10920838 | 0.27 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chrX_+_17755696 | 0.27 |
ENST00000419185.1
|
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr5_+_170288856 | 0.26 |
ENST00000523189.1
|
RANBP17
|
RAN binding protein 17 |
| chr17_+_34848049 | 0.26 |
ENST00000588902.1
ENST00000591067.1 |
ZNHIT3
|
zinc finger, HIT-type containing 3 |
| chr1_-_63782888 | 0.26 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr19_+_1452188 | 0.25 |
ENST00000587149.1
|
APC2
|
adenomatosis polyposis coli 2 |
| chr18_+_3247779 | 0.25 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr9_-_3469181 | 0.25 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr5_-_102455801 | 0.24 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr1_-_150669604 | 0.24 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr18_+_158513 | 0.24 |
ENST00000400266.3
ENST00000580410.1 ENST00000383589.2 ENST00000261601.7 |
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr11_+_74862032 | 0.24 |
ENST00000289575.5
ENST00000341411.4 |
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr15_+_63796779 | 0.23 |
ENST00000561442.1
ENST00000560070.1 ENST00000540797.1 ENST00000380324.3 ENST00000268049.7 ENST00000536001.1 ENST00000539772.1 |
USP3
|
ubiquitin specific peptidase 3 |
| chr12_+_104458235 | 0.23 |
ENST00000229330.4
|
HCFC2
|
host cell factor C2 |
| chr2_-_122494487 | 0.23 |
ENST00000451734.1
ENST00000285814.4 |
MKI67IP
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr10_+_94594351 | 0.23 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr8_+_33342268 | 0.22 |
ENST00000360128.6
|
MAK16
|
MAK16 homolog (S. cerevisiae) |
| chr9_+_6758109 | 0.22 |
ENST00000536108.1
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr17_+_38083977 | 0.22 |
ENST00000578802.1
ENST00000578478.1 ENST00000582263.1 |
RP11-387H17.4
|
RP11-387H17.4 |
| chr6_-_30080876 | 0.21 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr21_+_30673091 | 0.21 |
ENST00000447177.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr11_+_74862140 | 0.21 |
ENST00000525650.1
ENST00000454962.2 |
SLCO2B1
|
solute carrier organic anion transporter family, member 2B1 |
| chr3_+_169684553 | 0.21 |
ENST00000337002.4
ENST00000480708.1 |
SEC62
|
SEC62 homolog (S. cerevisiae) |
| chr17_-_191188 | 0.21 |
ENST00000575634.1
|
RPH3AL
|
rabphilin 3A-like (without C2 domains) |
| chr10_-_7661623 | 0.20 |
ENST00000298441.6
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5 |
| chr2_+_171627597 | 0.20 |
ENST00000429172.1
ENST00000426475.1 |
AC007405.6
|
AC007405.6 |
| chr12_+_56473628 | 0.20 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr14_-_53258180 | 0.20 |
ENST00000554230.1
|
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr13_-_50510434 | 0.19 |
ENST00000361840.3
|
SPRYD7
|
SPRY domain containing 7 |
| chr13_-_50510622 | 0.19 |
ENST00000378195.2
|
SPRYD7
|
SPRY domain containing 7 |
| chr3_+_122296465 | 0.19 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr8_-_82633476 | 0.19 |
ENST00000518419.1
ENST00000517588.1 ENST00000521895.1 ENST00000520076.1 ENST00000519523.1 ENST00000522520.1 ENST00000521287.1 ENST00000523096.1 ENST00000220669.5 ENST00000517450.1 |
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr1_-_114429997 | 0.19 |
ENST00000471267.1
ENST00000393320.3 |
BCL2L15
|
BCL2-like 15 |
| chr3_-_176915215 | 0.19 |
ENST00000457928.2
ENST00000422442.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr11_-_14913765 | 0.19 |
ENST00000334636.5
|
CYP2R1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr14_-_23288930 | 0.19 |
ENST00000554517.1
ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr17_+_39421591 | 0.19 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr9_+_6758024 | 0.19 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr14_-_50154921 | 0.19 |
ENST00000553805.2
ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2
|
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr1_+_78354297 | 0.18 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chr3_+_179065474 | 0.18 |
ENST00000471841.1
ENST00000280653.7 |
MFN1
|
mitofusin 1 |
| chr8_-_19540266 | 0.18 |
ENST00000311540.4
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr8_-_55014415 | 0.18 |
ENST00000522007.1
ENST00000521898.1 ENST00000518546.1 ENST00000316963.3 |
LYPLA1
|
lysophospholipase I |
| chr1_-_150849208 | 0.18 |
ENST00000358595.5
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr17_-_39183452 | 0.18 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr20_-_1165319 | 0.18 |
ENST00000429036.1
|
TMEM74B
|
transmembrane protein 74B |
| chr2_+_113321939 | 0.18 |
ENST00000458012.2
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr3_+_183948161 | 0.18 |
ENST00000426955.2
|
VWA5B2
|
von Willebrand factor A domain containing 5B2 |
| chr15_+_57998821 | 0.18 |
ENST00000299638.3
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr15_-_99548775 | 0.18 |
ENST00000378919.6
|
PGPEP1L
|
pyroglutamyl-peptidase I-like |
| chr12_+_72233487 | 0.18 |
ENST00000482439.2
ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15
|
TBC1 domain family, member 15 |
| chr6_+_31554962 | 0.18 |
ENST00000376092.3
ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1
|
leukocyte specific transcript 1 |
| chr6_+_44355257 | 0.17 |
ENST00000371477.3
|
CDC5L
|
cell division cycle 5-like |
| chrX_-_108868390 | 0.17 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr8_-_19540086 | 0.17 |
ENST00000332246.6
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr2_+_33701707 | 0.17 |
ENST00000425210.1
ENST00000444784.1 ENST00000423159.1 |
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr1_-_6550625 | 0.17 |
ENST00000377725.1
ENST00000340850.5 |
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr2_+_192110199 | 0.17 |
ENST00000304164.4
|
MYO1B
|
myosin IB |
| chr5_+_68513557 | 0.17 |
ENST00000256441.4
|
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr11_-_64512469 | 0.17 |
ENST00000377485.1
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr15_+_57998923 | 0.17 |
ENST00000380557.4
|
POLR2M
|
polymerase (RNA) II (DNA directed) polypeptide M |
| chr10_-_51130715 | 0.17 |
ENST00000402038.3
|
PARG
|
poly (ADP-ribose) glycohydrolase |
| chr5_-_58571935 | 0.16 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr3_-_182833863 | 0.16 |
ENST00000492597.1
|
MCCC1
|
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr15_+_89787180 | 0.16 |
ENST00000300027.8
ENST00000310775.7 ENST00000567891.1 ENST00000564920.1 ENST00000565255.1 ENST00000567996.1 ENST00000451393.2 ENST00000563250.1 |
FANCI
|
Fanconi anemia, complementation group I |
| chr16_+_55522536 | 0.16 |
ENST00000570283.1
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chr12_-_52845910 | 0.16 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr12_-_68845417 | 0.16 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr17_+_4487294 | 0.16 |
ENST00000338859.4
|
SMTNL2
|
smoothelin-like 2 |
| chr14_+_31091511 | 0.16 |
ENST00000544052.2
ENST00000421551.3 ENST00000541123.1 ENST00000557076.1 ENST00000553693.1 ENST00000396629.2 |
SCFD1
|
sec1 family domain containing 1 |
| chr4_+_95972822 | 0.16 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr12_+_50355647 | 0.16 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr9_-_100954910 | 0.16 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr12_+_16500599 | 0.16 |
ENST00000535309.1
ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr16_-_9030515 | 0.15 |
ENST00000535863.1
ENST00000381886.4 |
USP7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr17_-_7081435 | 0.15 |
ENST00000380920.4
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr1_-_39339777 | 0.15 |
ENST00000397572.2
|
MYCBP
|
MYC binding protein |
| chr2_-_158345341 | 0.15 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr2_+_68384976 | 0.15 |
ENST00000263657.2
|
PNO1
|
partner of NOB1 homolog (S. cerevisiae) |
| chr6_-_13711773 | 0.14 |
ENST00000011619.3
|
RANBP9
|
RAN binding protein 9 |
| chr6_+_143772060 | 0.14 |
ENST00000367591.4
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr12_+_16500571 | 0.14 |
ENST00000543076.1
ENST00000396210.3 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr6_-_27835357 | 0.14 |
ENST00000331442.3
|
HIST1H1B
|
histone cluster 1, H1b |
| chr5_-_142784101 | 0.14 |
ENST00000503201.1
ENST00000502892.1 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr10_+_17686221 | 0.14 |
ENST00000540523.1
|
STAM
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr10_-_12084770 | 0.14 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr19_-_45826125 | 0.14 |
ENST00000221476.3
|
CKM
|
creatine kinase, muscle |
| chr2_-_190627481 | 0.14 |
ENST00000264151.5
ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr20_-_23030296 | 0.14 |
ENST00000377103.2
|
THBD
|
thrombomodulin |
| chr7_+_99102573 | 0.14 |
ENST00000394170.2
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5 |
| chr9_-_130541017 | 0.14 |
ENST00000314830.8
|
SH2D3C
|
SH2 domain containing 3C |
| chr14_+_69658194 | 0.14 |
ENST00000409018.3
ENST00000409014.1 ENST00000409675.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chr8_-_30013748 | 0.13 |
ENST00000607315.1
|
RP11-51J9.5
|
RP11-51J9.5 |
| chr7_+_87563557 | 0.13 |
ENST00000439864.1
ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22
|
ADAM metallopeptidase domain 22 |
| chr5_+_49962772 | 0.13 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr12_+_8850471 | 0.13 |
ENST00000535829.1
ENST00000357529.3 |
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr3_-_10332416 | 0.13 |
ENST00000450603.1
ENST00000449554.2 |
GHRL
|
ghrelin/obestatin prepropeptide |
| chr4_-_89152474 | 0.13 |
ENST00000515655.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr13_+_37574678 | 0.13 |
ENST00000389704.3
|
EXOSC8
|
exosome component 8 |
| chr6_-_31648150 | 0.13 |
ENST00000375858.3
ENST00000383237.4 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr3_+_145782358 | 0.13 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr6_+_116832789 | 0.13 |
ENST00000368599.3
|
FAM26E
|
family with sequence similarity 26, member E |
| chr2_-_224702270 | 0.13 |
ENST00000396654.2
ENST00000396653.2 ENST00000423110.1 ENST00000443700.1 |
AP1S3
|
adaptor-related protein complex 1, sigma 3 subunit |
| chr19_-_50400212 | 0.13 |
ENST00000391826.2
|
IL4I1
|
interleukin 4 induced 1 |
| chr2_+_33701286 | 0.13 |
ENST00000403687.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chrX_-_16887963 | 0.13 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr20_-_1165117 | 0.13 |
ENST00000381894.3
|
TMEM74B
|
transmembrane protein 74B |
| chr10_-_46641003 | 0.13 |
ENST00000395721.2
ENST00000374218.2 ENST00000395725.3 ENST00000374346.3 ENST00000417004.1 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr12_-_105352080 | 0.13 |
ENST00000433540.1
|
SLC41A2
|
solute carrier family 41 (magnesium transporter), member 2 |
| chr4_-_123843597 | 0.13 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chr2_-_191115229 | 0.12 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr12_+_9142131 | 0.12 |
ENST00000356986.3
ENST00000266551.4 |
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr7_+_66205712 | 0.12 |
ENST00000451741.2
ENST00000442563.1 ENST00000450873.2 ENST00000284957.5 |
KCTD7
RABGEF1
|
potassium channel tetramerization domain containing 7 RAB guanine nucleotide exchange factor (GEF) 1 |
| chr14_-_45431091 | 0.12 |
ENST00000579157.1
ENST00000396128.4 ENST00000556500.1 |
KLHL28
|
kelch-like family member 28 |
| chr8_-_123706338 | 0.12 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr9_+_6757634 | 0.12 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr1_+_25757376 | 0.12 |
ENST00000399766.3
ENST00000399763.3 ENST00000374343.4 |
TMEM57
|
transmembrane protein 57 |
| chr3_-_54962100 | 0.12 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr11_-_88070920 | 0.12 |
ENST00000524463.1
ENST00000227266.5 |
CTSC
|
cathepsin C |
| chr2_+_234602305 | 0.12 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr11_-_82997420 | 0.12 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr3_+_197677379 | 0.12 |
ENST00000442341.1
|
RPL35A
|
ribosomal protein L35a |
| chr12_+_14572070 | 0.12 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr16_+_5009250 | 0.12 |
ENST00000587469.1
|
SEC14L5
|
SEC14-like 5 (S. cerevisiae) |
| chr14_+_58666824 | 0.12 |
ENST00000254286.4
|
ACTR10
|
actin-related protein 10 homolog (S. cerevisiae) |
| chr10_+_60094735 | 0.11 |
ENST00000373910.4
|
UBE2D1
|
ubiquitin-conjugating enzyme E2D 1 |
| chr16_-_832926 | 0.11 |
ENST00000293892.3
|
MSLNL
|
mesothelin-like |
| chr6_-_72130472 | 0.11 |
ENST00000426635.2
|
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr4_+_186347388 | 0.11 |
ENST00000511138.1
ENST00000511581.1 |
C4orf47
|
chromosome 4 open reading frame 47 |
| chr21_-_38362497 | 0.11 |
ENST00000427746.1
ENST00000336648.4 |
HLCS
|
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
| chr8_+_38089198 | 0.11 |
ENST00000528358.1
ENST00000529642.1 ENST00000532222.1 ENST00000520272.2 |
DDHD2
|
DDHD domain containing 2 |
| chr3_+_49059038 | 0.11 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr6_+_150070857 | 0.11 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr15_+_75487984 | 0.11 |
ENST00000563905.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr11_+_125496619 | 0.11 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr17_+_19314505 | 0.11 |
ENST00000461366.1
|
RNF112
|
ring finger protein 112 |
| chr19_-_4559814 | 0.11 |
ENST00000586582.1
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr5_-_98262240 | 0.11 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr17_-_26989136 | 0.11 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr7_+_12726623 | 0.10 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chrX_+_9880412 | 0.10 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr6_+_150070831 | 0.10 |
ENST00000367380.5
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr7_-_25164868 | 0.10 |
ENST00000409409.1
ENST00000409764.1 ENST00000413447.1 |
CYCS
|
cytochrome c, somatic |
| chr3_+_113666748 | 0.10 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr16_-_3422283 | 0.10 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr19_+_57999101 | 0.10 |
ENST00000347466.6
ENST00000523138.1 ENST00000415379.2 ENST00000521754.1 ENST00000221735.7 ENST00000518999.1 ENST00000521137.1 |
ZNF419
|
zinc finger protein 419 |
| chr6_+_31555045 | 0.10 |
ENST00000396101.3
ENST00000490742.1 |
LST1
|
leukocyte specific transcript 1 |
| chr14_+_67999999 | 0.10 |
ENST00000329153.5
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr4_-_140223670 | 0.10 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr3_+_49058444 | 0.10 |
ENST00000326925.6
ENST00000395458.2 |
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr7_-_48068643 | 0.10 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr7_-_132766800 | 0.10 |
ENST00000542753.1
ENST00000448878.1 |
CHCHD3
|
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr5_+_130506629 | 0.10 |
ENST00000510516.1
ENST00000507584.1 |
LYRM7
|
LYR motif containing 7 |
| chr1_-_150849174 | 0.10 |
ENST00000515192.1
|
ARNT
|
aryl hydrocarbon receptor nuclear translocator |
| chr6_+_17102489 | 0.09 |
ENST00000536551.1
|
STMND1
|
stathmin domain containing 1 |
| chr22_-_50964558 | 0.09 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr11_-_31531121 | 0.09 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF21
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 1.5 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.2 | 0.6 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.1 | 0.5 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.3 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.3 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.3 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.1 | 0.4 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.3 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.3 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.3 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:2000672 | cellular response to sorbitol(GO:0072709) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:1904344 | positive regulation of growth rate(GO:0040010) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.4 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.3 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.0 | 0.4 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.2 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.3 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.0 | 0.3 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.4 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.0 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.1 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 | 0.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.0 | 0.0 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.0 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.3 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.0 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.0 | 0.0 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0035932 | mineralocorticoid secretion(GO:0035931) aldosterone secretion(GO:0035932) regulation of mineralocorticoid secretion(GO:2000855) positive regulation of mineralocorticoid secretion(GO:2000857) regulation of aldosterone secretion(GO:2000858) positive regulation of aldosterone secretion(GO:2000860) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.0 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0034678 | smooth muscle contractile fiber(GO:0030485) integrin alpha8-beta1 complex(GO:0034678) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.5 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.3 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.2 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.1 | 0.3 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.1 | 0.3 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.4 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.1 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.8 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 3.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.0 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.0 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |