Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
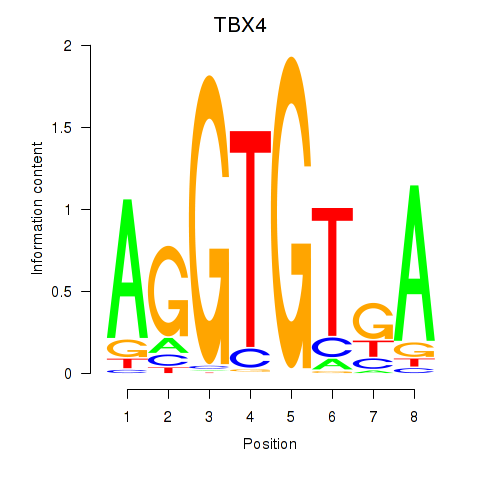
Results for TBX4
Z-value: 1.02
Transcription factors associated with TBX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX4
|
ENSG00000121075.5 | T-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX4 | hg19_v2_chr17_+_59529743_59529798 | 0.18 | 7.4e-01 | Click! |
Activity profile of TBX4 motif
Sorted Z-values of TBX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_167453493 | 1.51 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr2_-_61697862 | 0.80 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr3_+_167453026 | 0.74 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr8_-_99954788 | 0.70 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chrM_+_10464 | 0.64 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr5_+_74807581 | 0.63 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr7_+_107220660 | 0.61 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr21_+_30502806 | 0.55 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr14_-_62217779 | 0.54 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr5_+_74807886 | 0.54 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr2_-_111291587 | 0.47 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr9_+_40028620 | 0.46 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chrM_+_10758 | 0.46 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr8_-_95907423 | 0.44 |
ENST00000396133.3
ENST00000308108.4 |
CCNE2
|
cyclin E2 |
| chr7_+_107220899 | 0.42 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr2_-_37458749 | 0.36 |
ENST00000234170.5
|
CEBPZ
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr12_-_90024360 | 0.35 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr14_-_45603657 | 0.35 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr2_+_61108650 | 0.34 |
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr14_+_53196872 | 0.34 |
ENST00000442123.2
ENST00000354586.4 |
STYX
|
serine/threonine/tyrosine interacting protein |
| chr5_+_56469843 | 0.33 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chrM_+_10053 | 0.33 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr4_+_140222609 | 0.32 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr2_-_219925189 | 0.32 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr2_+_62933001 | 0.31 |
ENST00000263991.5
ENST00000354487.3 |
EHBP1
|
EH domain binding protein 1 |
| chr10_-_16563870 | 0.31 |
ENST00000298943.3
|
C1QL3
|
complement component 1, q subcomponent-like 3 |
| chr13_-_52027134 | 0.31 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr2_-_42160486 | 0.30 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr5_-_78809950 | 0.30 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chrX_+_123095546 | 0.30 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr1_+_16062820 | 0.30 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr11_-_85430204 | 0.29 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr7_-_105319536 | 0.29 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr21_+_30503282 | 0.29 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr11_+_111749650 | 0.28 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr5_-_60240858 | 0.28 |
ENST00000426742.2
ENST00000265038.5 ENST00000543101.1 ENST00000439176.1 |
ERCC8
|
excision repair cross-complementing rodent repair deficiency, complementation group 8 |
| chr14_+_75745477 | 0.28 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr3_+_142315294 | 0.28 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr3_+_142315225 | 0.27 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chrX_+_16804544 | 0.27 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr11_+_114271251 | 0.27 |
ENST00000375490.5
|
RBM7
|
RNA binding motif protein 7 |
| chr11_-_85779971 | 0.27 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr15_+_90319557 | 0.27 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr2_+_62932779 | 0.27 |
ENST00000427809.1
ENST00000405482.1 ENST00000431489.1 |
EHBP1
|
EH domain binding protein 1 |
| chrX_+_123095860 | 0.27 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chrX_-_133930285 | 0.26 |
ENST00000486347.1
ENST00000343004.5 |
FAM122B
|
family with sequence similarity 122B |
| chr11_-_85430163 | 0.26 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chrX_+_123095890 | 0.25 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr14_+_90864504 | 0.25 |
ENST00000544280.2
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr11_+_111750206 | 0.25 |
ENST00000530214.1
ENST00000530799.1 |
C11orf1
|
chromosome 11 open reading frame 1 |
| chr5_-_74807418 | 0.25 |
ENST00000405807.4
ENST00000261415.7 |
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr2_+_154728426 | 0.24 |
ENST00000392825.3
ENST00000434213.1 |
GALNT13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) |
| chr11_-_85430088 | 0.24 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr5_+_56469939 | 0.23 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr6_-_19804973 | 0.23 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr2_+_29033682 | 0.23 |
ENST00000379579.4
ENST00000334056.5 ENST00000449210.1 |
SPDYA
|
speedy/RINGO cell cycle regulator family member A |
| chr1_+_35734616 | 0.23 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr8_+_103563792 | 0.23 |
ENST00000285402.3
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr2_+_61108771 | 0.23 |
ENST00000394479.3
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr5_+_68788594 | 0.23 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr14_+_85994943 | 0.23 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr9_-_75567962 | 0.23 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr18_-_59561417 | 0.22 |
ENST00000591306.1
|
RNF152
|
ring finger protein 152 |
| chrX_+_69642881 | 0.22 |
ENST00000453994.2
ENST00000536730.1 ENST00000538649.1 ENST00000374382.3 |
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr7_-_28220354 | 0.22 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr5_+_82767487 | 0.21 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr5_+_56469775 | 0.20 |
ENST00000424459.3
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr4_-_146859787 | 0.19 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr11_+_4116054 | 0.19 |
ENST00000423050.2
|
RRM1
|
ribonucleotide reductase M1 |
| chr2_-_85895295 | 0.18 |
ENST00000428225.1
ENST00000519937.2 |
SFTPB
|
surfactant protein B |
| chrX_+_46772065 | 0.18 |
ENST00000455411.1
|
PHF16
|
jade family PHD finger 3 |
| chr12_+_104324112 | 0.18 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr1_-_1009683 | 0.18 |
ENST00000453464.2
|
RNF223
|
ring finger protein 223 |
| chr2_-_160473114 | 0.17 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr17_-_74533734 | 0.17 |
ENST00000589342.1
|
CYGB
|
cytoglobin |
| chr17_-_39203519 | 0.17 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr3_-_49459865 | 0.17 |
ENST00000427987.1
|
AMT
|
aminomethyltransferase |
| chr2_+_43864387 | 0.16 |
ENST00000282406.4
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr10_+_31608054 | 0.16 |
ENST00000320985.10
ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr4_-_114682719 | 0.16 |
ENST00000394522.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr7_-_150675372 | 0.16 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr5_+_60240943 | 0.16 |
ENST00000296597.5
|
NDUFAF2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr3_-_49459878 | 0.15 |
ENST00000546031.1
ENST00000458307.2 ENST00000430521.1 |
AMT
|
aminomethyltransferase |
| chr20_+_2517253 | 0.15 |
ENST00000358864.1
|
TMC2
|
transmembrane channel-like 2 |
| chr17_-_39222131 | 0.15 |
ENST00000394015.2
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr2_+_202937972 | 0.15 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr11_+_73358690 | 0.15 |
ENST00000545798.1
ENST00000539157.1 ENST00000546251.1 ENST00000535582.1 ENST00000538227.1 ENST00000543524.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr10_-_98945264 | 0.15 |
ENST00000314867.5
|
SLIT1
|
slit homolog 1 (Drosophila) |
| chr11_-_111749767 | 0.14 |
ENST00000542429.1
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr3_+_107364683 | 0.14 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr3_+_107364769 | 0.14 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr11_-_85430356 | 0.14 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr5_-_114515734 | 0.14 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr2_+_37458928 | 0.14 |
ENST00000439218.1
ENST00000432075.1 |
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr7_-_127032741 | 0.14 |
ENST00000393313.1
ENST00000265827.3 ENST00000434602.1 |
ZNF800
|
zinc finger protein 800 |
| chr17_+_30771279 | 0.14 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr11_-_114271139 | 0.14 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr11_+_111749942 | 0.14 |
ENST00000260276.3
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr6_-_130536774 | 0.14 |
ENST00000532763.1
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr2_+_103089756 | 0.14 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr15_+_76030311 | 0.14 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr15_-_83474806 | 0.13 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr11_+_18343800 | 0.13 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr14_+_75746781 | 0.13 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr5_+_82767284 | 0.13 |
ENST00000265077.3
|
VCAN
|
versican |
| chr2_+_37458776 | 0.13 |
ENST00000002125.4
ENST00000336237.6 ENST00000431821.1 |
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr11_-_18343669 | 0.12 |
ENST00000396253.3
ENST00000349215.3 ENST00000438420.2 |
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr11_+_74660278 | 0.12 |
ENST00000263672.6
ENST00000530257.1 ENST00000526361.1 ENST00000532972.1 |
SPCS2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr20_+_11898507 | 0.12 |
ENST00000378226.2
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chrX_+_46771848 | 0.11 |
ENST00000218343.4
|
PHF16
|
jade family PHD finger 3 |
| chr2_+_37458904 | 0.11 |
ENST00000416653.1
|
NDUFAF7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr14_+_75746664 | 0.11 |
ENST00000557139.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr11_+_18344106 | 0.11 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr9_+_140032842 | 0.10 |
ENST00000371561.3
ENST00000315048.3 |
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1 |
| chr17_+_57970469 | 0.10 |
ENST00000443572.2
ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| chr6_-_31080336 | 0.10 |
ENST00000259870.3
|
C6orf15
|
chromosome 6 open reading frame 15 |
| chr15_+_41913690 | 0.10 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr1_-_226076843 | 0.10 |
ENST00000272134.5
|
LEFTY1
|
left-right determination factor 1 |
| chr1_+_27114418 | 0.10 |
ENST00000078527.4
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr4_+_102268904 | 0.09 |
ENST00000527564.1
ENST00000529296.1 |
AP001816.1
|
Uncharacterized protein |
| chr1_+_95582881 | 0.09 |
ENST00000370203.4
ENST00000456991.1 |
TMEM56
|
transmembrane protein 56 |
| chr13_-_46543805 | 0.09 |
ENST00000378921.2
|
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr5_+_60241020 | 0.09 |
ENST00000511107.1
ENST00000502658.1 |
NDUFAF2
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr1_-_111148241 | 0.08 |
ENST00000440270.1
|
KCNA2
|
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr2_-_160472952 | 0.08 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr6_-_19804877 | 0.08 |
ENST00000447250.1
|
RP4-625H18.2
|
RP4-625H18.2 |
| chr1_-_37499726 | 0.08 |
ENST00000373091.3
ENST00000373093.4 |
GRIK3
|
glutamate receptor, ionotropic, kainate 3 |
| chr17_+_35849937 | 0.08 |
ENST00000394389.4
|
DUSP14
|
dual specificity phosphatase 14 |
| chr2_-_40680578 | 0.08 |
ENST00000455476.1
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr3_+_134514093 | 0.07 |
ENST00000398015.3
|
EPHB1
|
EPH receptor B1 |
| chr3_+_120626919 | 0.07 |
ENST00000273666.6
ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L
|
syntaxin binding protein 5-like |
| chr15_+_76016293 | 0.07 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr5_-_34043310 | 0.07 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr6_+_39760129 | 0.07 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr11_+_114270752 | 0.07 |
ENST00000540163.1
|
RBM7
|
RNA binding motif protein 7 |
| chr8_+_56014949 | 0.07 |
ENST00000327381.6
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4 |
| chr7_+_133261209 | 0.06 |
ENST00000545148.1
|
EXOC4
|
exocyst complex component 4 |
| chr11_-_125932685 | 0.06 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr22_-_41215291 | 0.06 |
ENST00000542412.1
ENST00000544408.1 |
SLC25A17
|
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
| chrX_+_70443050 | 0.06 |
ENST00000361726.6
|
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr6_+_30295036 | 0.06 |
ENST00000376659.5
ENST00000428555.1 |
TRIM39
|
tripartite motif containing 39 |
| chr11_+_17568920 | 0.05 |
ENST00000399391.2
|
OTOG
|
otogelin |
| chr6_+_79577189 | 0.05 |
ENST00000369940.2
|
IRAK1BP1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr3_-_197024394 | 0.04 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr3_+_52422899 | 0.04 |
ENST00000480649.1
|
DNAH1
|
dynein, axonemal, heavy chain 1 |
| chr2_-_61108449 | 0.04 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr12_+_58148842 | 0.04 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr3_-_133748913 | 0.04 |
ENST00000310926.4
|
SLCO2A1
|
solute carrier organic anion transporter family, member 2A1 |
| chr2_+_171571827 | 0.04 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr3_+_63805017 | 0.04 |
ENST00000295896.8
|
C3orf49
|
chromosome 3 open reading frame 49 |
| chrX_+_46771711 | 0.04 |
ENST00000424392.1
ENST00000397189.1 |
PHF16
|
jade family PHD finger 3 |
| chr11_+_86511549 | 0.04 |
ENST00000533902.2
|
PRSS23
|
protease, serine, 23 |
| chr14_+_21467414 | 0.03 |
ENST00000554422.1
ENST00000298681.4 |
SLC39A2
|
solute carrier family 39 (zinc transporter), member 2 |
| chr7_-_71801980 | 0.03 |
ENST00000329008.5
|
CALN1
|
calneuron 1 |
| chr2_+_219081817 | 0.03 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr9_-_74979420 | 0.03 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr1_+_172628154 | 0.03 |
ENST00000340030.3
ENST00000367721.2 |
FASLG
|
Fas ligand (TNF superfamily, member 6) |
| chr20_+_44657845 | 0.03 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr4_-_140222358 | 0.03 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr15_+_92937058 | 0.03 |
ENST00000268164.3
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr15_+_36994210 | 0.03 |
ENST00000562489.1
|
C15orf41
|
chromosome 15 open reading frame 41 |
| chr1_+_206680879 | 0.03 |
ENST00000355294.4
ENST00000367117.3 |
RASSF5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr11_-_34535332 | 0.03 |
ENST00000257832.2
ENST00000429939.2 |
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr4_-_102268484 | 0.02 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr13_-_103053946 | 0.02 |
ENST00000376131.4
|
FGF14
|
fibroblast growth factor 14 |
| chr11_-_128737163 | 0.02 |
ENST00000324003.3
ENST00000392665.2 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr4_-_186456652 | 0.02 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr15_-_38852251 | 0.02 |
ENST00000558432.1
|
RASGRP1
|
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr6_+_32132360 | 0.02 |
ENST00000333845.6
ENST00000395512.1 ENST00000432129.1 |
EGFL8
|
EGF-like-domain, multiple 8 |
| chr4_-_114682597 | 0.02 |
ENST00000394524.3
|
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
| chr15_-_58357932 | 0.02 |
ENST00000347587.3
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2 |
| chr13_-_36429763 | 0.02 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr10_+_135050908 | 0.02 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr11_+_122709200 | 0.02 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr12_-_26278030 | 0.01 |
ENST00000242728.4
|
BHLHE41
|
basic helix-loop-helix family, member e41 |
| chr6_+_30294612 | 0.01 |
ENST00000440271.1
ENST00000396551.3 ENST00000376656.4 ENST00000540416.1 ENST00000428728.1 ENST00000396548.1 ENST00000428404.1 |
TRIM39
|
tripartite motif containing 39 |
| chr1_+_26737292 | 0.01 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr2_+_176972000 | 0.01 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr2_-_175869936 | 0.01 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr11_-_34535297 | 0.01 |
ENST00000532417.1
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr11_-_33913708 | 0.01 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr17_-_37557846 | 0.01 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr4_-_186456766 | 0.01 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr15_+_92937144 | 0.01 |
ENST00000539113.1
ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr20_+_44657807 | 0.01 |
ENST00000372315.1
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr18_-_53068911 | 0.00 |
ENST00000537856.3
|
TCF4
|
transcription factor 4 |
| chr4_-_114682936 | 0.00 |
ENST00000454265.2
ENST00000429180.1 ENST00000418639.2 ENST00000394526.2 ENST00000296402.5 |
CAMK2D
|
calcium/calmodulin-dependent protein kinase II delta |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.3 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 1.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.2 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.9 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.8 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.3 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.4 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.1 | 0.2 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.4 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 1.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.6 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.7 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 2.2 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.0 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.0 | 0.0 | GO:0043132 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.0 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 1.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 0.4 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 1.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 1.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.0 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |