Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
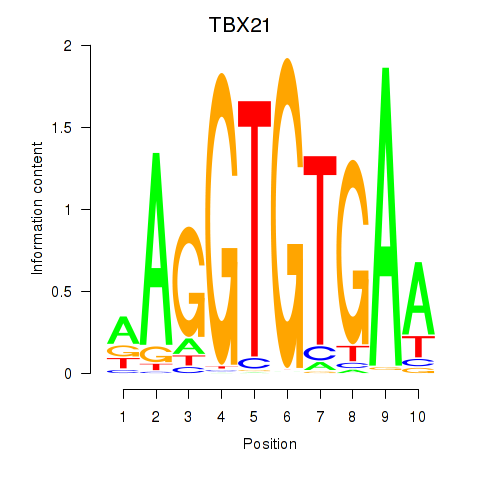
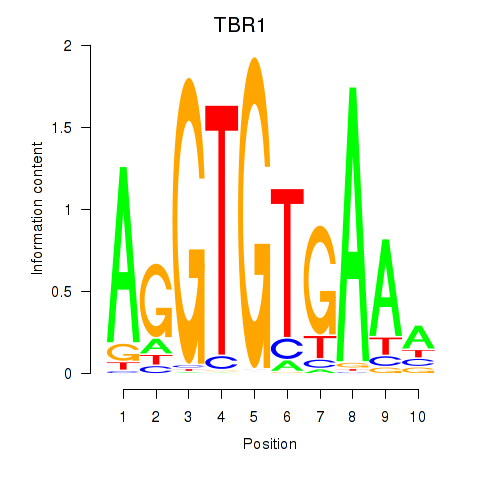
Results for TBX21_TBR1
Z-value: 0.44
Transcription factors associated with TBX21_TBR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX21
|
ENSG00000073861.2 | T-box transcription factor 21 |
|
TBR1
|
ENSG00000136535.10 | T-box brain transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBR1 | hg19_v2_chr2_+_162272605_162272753 | 0.34 | 5.2e-01 | Click! |
Activity profile of TBX21_TBR1 motif
Sorted Z-values of TBX21_TBR1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_29001711 | 0.33 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr10_+_135050908 | 0.30 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr14_-_62217779 | 0.25 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr15_-_83316254 | 0.23 |
ENST00000567678.1
ENST00000450751.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr7_+_107220660 | 0.21 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr11_-_114271139 | 0.19 |
ENST00000325636.4
|
C11orf71
|
chromosome 11 open reading frame 71 |
| chr19_-_35068562 | 0.18 |
ENST00000595013.1
|
SCGB1B2P
|
secretoglobin, family 1B, member 2, pseudogene |
| chr17_-_34122596 | 0.17 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr5_-_131132614 | 0.17 |
ENST00000307968.7
ENST00000307954.8 |
FNIP1
|
folliculin interacting protein 1 |
| chr15_+_90319557 | 0.16 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chr22_-_31688381 | 0.16 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr14_+_32546145 | 0.16 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_-_205744205 | 0.16 |
ENST00000446390.2
|
RAB7L1
|
RAB7, member RAS oncogene family-like 1 |
| chr11_-_133826852 | 0.15 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr4_+_124317940 | 0.15 |
ENST00000505319.1
ENST00000339241.1 |
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr16_+_12059091 | 0.15 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr15_-_83315874 | 0.15 |
ENST00000569257.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr12_-_68845417 | 0.15 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr4_-_177116772 | 0.15 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr15_-_80695917 | 0.14 |
ENST00000559008.1
|
RP11-210M15.2
|
Uncharacterized protein |
| chr3_-_197024394 | 0.14 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr11_+_122709200 | 0.14 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr2_+_87808725 | 0.14 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr14_+_32546274 | 0.14 |
ENST00000396582.2
|
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr1_+_16062820 | 0.14 |
ENST00000294454.5
|
SLC25A34
|
solute carrier family 25, member 34 |
| chr17_+_76356516 | 0.13 |
ENST00000592569.1
|
RP11-806H10.4
|
RP11-806H10.4 |
| chr2_+_85646054 | 0.13 |
ENST00000389938.2
|
SH2D6
|
SH2 domain containing 6 |
| chr16_-_67450325 | 0.13 |
ENST00000348579.2
|
ZDHHC1
|
zinc finger, DHHC-type containing 1 |
| chr11_+_82783097 | 0.13 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr1_-_1009683 | 0.12 |
ENST00000453464.2
|
RNF223
|
ring finger protein 223 |
| chr11_-_82782861 | 0.12 |
ENST00000524635.1
ENST00000526205.1 ENST00000527633.1 ENST00000533486.1 ENST00000533276.2 |
RAB30
|
RAB30, member RAS oncogene family |
| chr4_-_185303418 | 0.12 |
ENST00000610223.1
ENST00000608785.1 |
RP11-290F5.1
|
RP11-290F5.1 |
| chr9_+_26956371 | 0.12 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr1_+_244227632 | 0.12 |
ENST00000598000.1
|
AL590483.1
|
Uncharacterized protein; cDNA FLJ42623 fis, clone BRACE3015894 |
| chr6_-_105584560 | 0.11 |
ENST00000336775.5
|
BVES
|
blood vessel epicardial substance |
| chr14_+_85994943 | 0.11 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr11_+_64073699 | 0.11 |
ENST00000405666.1
ENST00000468670.1 |
ESRRA
|
estrogen-related receptor alpha |
| chr7_-_127032741 | 0.11 |
ENST00000393313.1
ENST00000265827.3 ENST00000434602.1 |
ZNF800
|
zinc finger protein 800 |
| chr6_-_84937314 | 0.11 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr1_+_28586006 | 0.11 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr2_-_61697862 | 0.10 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr5_-_131132658 | 0.10 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr3_+_112709804 | 0.10 |
ENST00000383677.3
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr11_+_65266507 | 0.10 |
ENST00000544868.1
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr19_+_3762703 | 0.10 |
ENST00000589174.1
|
MRPL54
|
mitochondrial ribosomal protein L54 |
| chr22_+_18593097 | 0.10 |
ENST00000426208.1
|
TUBA8
|
tubulin, alpha 8 |
| chr1_-_163172625 | 0.10 |
ENST00000527988.1
ENST00000531476.1 ENST00000530507.1 |
RGS5
|
regulator of G-protein signaling 5 |
| chr17_-_39150385 | 0.10 |
ENST00000391586.1
|
KRTAP3-3
|
keratin associated protein 3-3 |
| chr2_+_102758751 | 0.10 |
ENST00000442590.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr16_-_11350036 | 0.10 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr10_-_76868931 | 0.09 |
ENST00000372700.3
ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13
|
dual specificity phosphatase 13 |
| chr12_+_14572070 | 0.09 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr7_+_107220899 | 0.09 |
ENST00000379117.2
ENST00000473124.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr14_-_65569244 | 0.09 |
ENST00000557277.1
ENST00000556892.1 |
MAX
|
MYC associated factor X |
| chr19_+_36236514 | 0.09 |
ENST00000222266.2
|
PSENEN
|
presenilin enhancer gamma secretase subunit |
| chr16_+_75690093 | 0.09 |
ENST00000564671.2
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein |
| chr1_+_156308245 | 0.09 |
ENST00000368253.2
ENST00000470342.1 ENST00000368254.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr5_+_140571902 | 0.09 |
ENST00000239446.4
|
PCDHB10
|
protocadherin beta 10 |
| chr6_-_97731019 | 0.09 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr14_-_45603657 | 0.09 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr9_+_35829208 | 0.09 |
ENST00000439587.2
ENST00000377991.4 |
TMEM8B
|
transmembrane protein 8B |
| chr18_-_59561417 | 0.09 |
ENST00000591306.1
|
RNF152
|
ring finger protein 152 |
| chr5_+_132009675 | 0.09 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr8_+_145490549 | 0.08 |
ENST00000340695.2
|
SCXA
|
scleraxis homolog A (mouse) |
| chrX_+_99899180 | 0.08 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr20_+_2517253 | 0.08 |
ENST00000358864.1
|
TMC2
|
transmembrane channel-like 2 |
| chr12_-_110511424 | 0.08 |
ENST00000548191.1
|
C12orf76
|
chromosome 12 open reading frame 76 |
| chr11_+_111412271 | 0.08 |
ENST00000528102.1
|
LAYN
|
layilin |
| chr5_+_74807886 | 0.08 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr1_+_100111580 | 0.08 |
ENST00000605497.1
|
PALMD
|
palmdelphin |
| chr11_+_107799118 | 0.08 |
ENST00000320578.2
|
RAB39A
|
RAB39A, member RAS oncogene family |
| chr12_+_66582919 | 0.08 |
ENST00000545837.1
ENST00000457197.2 |
IRAK3
|
interleukin-1 receptor-associated kinase 3 |
| chr16_-_66968055 | 0.08 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr20_+_44098385 | 0.08 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr17_+_6659354 | 0.08 |
ENST00000574907.1
|
XAF1
|
XIAP associated factor 1 |
| chr8_-_61880248 | 0.08 |
ENST00000525556.1
|
AC022182.3
|
AC022182.3 |
| chr1_+_156105878 | 0.08 |
ENST00000508500.1
|
LMNA
|
lamin A/C |
| chr3_-_196065248 | 0.08 |
ENST00000446879.1
ENST00000273695.3 |
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr1_+_174843548 | 0.08 |
ENST00000478442.1
ENST00000465412.1 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr1_+_38512799 | 0.08 |
ENST00000432922.1
ENST00000428151.1 |
RP5-884C9.2
|
RP5-884C9.2 |
| chr1_-_44482979 | 0.08 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr22_+_23243156 | 0.08 |
ENST00000390323.2
|
IGLC2
|
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr19_-_13213662 | 0.08 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr1_+_100111479 | 0.07 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chr1_-_76076759 | 0.07 |
ENST00000370855.5
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr17_+_4046964 | 0.07 |
ENST00000573984.1
|
CYB5D2
|
cytochrome b5 domain containing 2 |
| chr9_-_35815013 | 0.07 |
ENST00000259667.5
|
HINT2
|
histidine triad nucleotide binding protein 2 |
| chr17_-_42452063 | 0.07 |
ENST00000588098.1
|
ITGA2B
|
integrin, alpha 2b (platelet glycoprotein IIb of IIb/IIIa complex, antigen CD41) |
| chr8_-_99954788 | 0.07 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr11_-_82782952 | 0.07 |
ENST00000534141.1
|
RAB30
|
RAB30, member RAS oncogene family |
| chr17_+_42925270 | 0.07 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr17_-_1553346 | 0.07 |
ENST00000301336.6
|
RILP
|
Rab interacting lysosomal protein |
| chr2_+_207139367 | 0.07 |
ENST00000374423.3
|
ZDBF2
|
zinc finger, DBF-type containing 2 |
| chr2_-_61245363 | 0.07 |
ENST00000316752.6
|
PUS10
|
pseudouridylate synthase 10 |
| chr8_+_103563792 | 0.07 |
ENST00000285402.3
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr2_+_102758271 | 0.07 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr2_+_233404429 | 0.07 |
ENST00000389494.3
ENST00000389492.3 |
CHRNG
|
cholinergic receptor, nicotinic, gamma (muscle) |
| chr10_+_24497704 | 0.07 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr13_+_113548643 | 0.07 |
ENST00000375608.3
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr2_+_152266392 | 0.07 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr6_-_26124138 | 0.07 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr1_-_26633067 | 0.06 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr2_-_64751227 | 0.06 |
ENST00000561559.1
|
RP11-568N6.1
|
RP11-568N6.1 |
| chr2_+_120687335 | 0.06 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr10_-_126432821 | 0.06 |
ENST00000280780.6
|
FAM53B
|
family with sequence similarity 53, member B |
| chr15_-_83316087 | 0.06 |
ENST00000568757.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr1_-_44818599 | 0.06 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr7_+_110731062 | 0.06 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr1_+_213123976 | 0.06 |
ENST00000366965.2
ENST00000366967.2 |
VASH2
|
vasohibin 2 |
| chr14_-_20881579 | 0.06 |
ENST00000556935.1
ENST00000262715.5 ENST00000556549.1 |
TEP1
|
telomerase-associated protein 1 |
| chr10_-_23003460 | 0.06 |
ENST00000376573.4
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr1_+_89829610 | 0.06 |
ENST00000370456.4
ENST00000535065.1 |
GBP6
|
guanylate binding protein family, member 6 |
| chr17_+_35849937 | 0.06 |
ENST00000394389.4
|
DUSP14
|
dual specificity phosphatase 14 |
| chr4_-_89619386 | 0.06 |
ENST00000323061.5
|
NAP1L5
|
nucleosome assembly protein 1-like 5 |
| chr8_-_30890710 | 0.06 |
ENST00000523392.1
|
PURG
|
purine-rich element binding protein G |
| chr4_-_121843985 | 0.06 |
ENST00000264808.3
ENST00000428209.2 ENST00000515109.1 ENST00000394435.2 |
PRDM5
|
PR domain containing 5 |
| chr1_-_100643765 | 0.06 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr3_-_61237050 | 0.06 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr3_+_112709755 | 0.06 |
ENST00000383678.2
|
GTPBP8
|
GTP-binding protein 8 (putative) |
| chr2_+_228736335 | 0.06 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr2_-_228497888 | 0.06 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr22_+_31003133 | 0.06 |
ENST00000405742.3
|
TCN2
|
transcobalamin II |
| chr21_+_25801041 | 0.06 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr17_-_47308128 | 0.06 |
ENST00000413580.1
ENST00000511066.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr14_-_24584138 | 0.06 |
ENST00000558280.1
ENST00000561028.1 |
NRL
|
neural retina leucine zipper |
| chr18_-_48723129 | 0.06 |
ENST00000592416.1
|
MEX3C
|
mex-3 RNA binding family member C |
| chr17_-_47308100 | 0.05 |
ENST00000503902.1
ENST00000512250.1 |
PHOSPHO1
|
phosphatase, orphan 1 |
| chr5_+_74807581 | 0.05 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr14_-_98444438 | 0.05 |
ENST00000512901.2
|
C14orf64
|
chromosome 14 open reading frame 64 |
| chr4_-_186732048 | 0.05 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr1_-_113247543 | 0.05 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr1_-_76076793 | 0.05 |
ENST00000370859.3
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr6_+_108616054 | 0.05 |
ENST00000368977.4
|
LACE1
|
lactation elevated 1 |
| chr15_+_76030311 | 0.05 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr11_+_17756279 | 0.05 |
ENST00000265969.6
|
KCNC1
|
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chr20_+_30225682 | 0.05 |
ENST00000376075.3
|
COX4I2
|
cytochrome c oxidase subunit IV isoform 2 (lung) |
| chr12_-_113573495 | 0.05 |
ENST00000446861.3
|
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr9_+_125273081 | 0.05 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor, family 1, subfamily J, member 2 |
| chr6_-_13487784 | 0.05 |
ENST00000379287.3
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr16_+_53133070 | 0.05 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr1_+_156308403 | 0.05 |
ENST00000481479.1
ENST00000368252.1 ENST00000466306.1 ENST00000368251.1 |
TSACC
|
TSSK6 activating co-chaperone |
| chr22_+_18560743 | 0.05 |
ENST00000399744.3
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr20_+_44098346 | 0.05 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr15_-_81195510 | 0.05 |
ENST00000561295.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr12_+_57854274 | 0.05 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr1_-_115124257 | 0.05 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr18_-_30050395 | 0.05 |
ENST00000269209.6
ENST00000399218.4 |
GAREM
|
GRB2 associated, regulator of MAPK1 |
| chr1_+_212782012 | 0.05 |
ENST00000341491.4
ENST00000366985.1 |
ATF3
|
activating transcription factor 3 |
| chr22_+_31003190 | 0.05 |
ENST00000407817.3
|
TCN2
|
transcobalamin II |
| chr22_+_18560675 | 0.05 |
ENST00000329627.7
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chrX_+_69642881 | 0.05 |
ENST00000453994.2
ENST00000536730.1 ENST00000538649.1 ENST00000374382.3 |
GDPD2
|
glycerophosphodiester phosphodiesterase domain containing 2 |
| chr2_+_219246746 | 0.05 |
ENST00000233202.6
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr5_-_74807418 | 0.05 |
ENST00000405807.4
ENST00000261415.7 |
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr6_-_130536774 | 0.05 |
ENST00000532763.1
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr16_+_72459838 | 0.05 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr11_+_129685707 | 0.05 |
ENST00000281441.3
|
TMEM45B
|
transmembrane protein 45B |
| chr12_+_69742121 | 0.05 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr5_+_6448736 | 0.05 |
ENST00000399816.3
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chrX_-_106146547 | 0.05 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr12_-_76879852 | 0.05 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr12_-_71148413 | 0.05 |
ENST00000440835.2
ENST00000549308.1 ENST00000550661.1 |
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr7_-_91875358 | 0.05 |
ENST00000458177.1
ENST00000394507.1 ENST00000340022.2 ENST00000444960.1 |
KRIT1
|
KRIT1, ankyrin repeat containing |
| chr16_+_50313426 | 0.05 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr16_-_18937726 | 0.05 |
ENST00000389467.3
ENST00000446231.2 |
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr9_-_132906498 | 0.05 |
ENST00000358748.1
|
AL360004.1
|
Uncharacterized protein |
| chr2_+_102758210 | 0.05 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr9_+_22646189 | 0.05 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr18_-_6928495 | 0.05 |
ENST00000580197.1
|
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr9_+_5510558 | 0.05 |
ENST00000397747.3
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr19_+_6373715 | 0.05 |
ENST00000599849.1
|
ALKBH7
|
alkB, alkylation repair homolog 7 (E. coli) |
| chr19_+_56653064 | 0.05 |
ENST00000593100.1
|
ZNF444
|
zinc finger protein 444 |
| chr17_-_7493390 | 0.05 |
ENST00000538513.2
ENST00000570788.1 ENST00000250055.2 |
SOX15
|
SRY (sex determining region Y)-box 15 |
| chr1_+_111682827 | 0.05 |
ENST00000357172.4
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr17_+_54911444 | 0.05 |
ENST00000284061.3
ENST00000572810.1 |
DGKE
|
diacylglycerol kinase, epsilon 64kDa |
| chr17_-_46657473 | 0.05 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr15_+_59908633 | 0.04 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr12_-_113574028 | 0.04 |
ENST00000546530.1
ENST00000261729.5 |
RASAL1
|
RAS protein activator like 1 (GAP1 like) |
| chr6_+_45390222 | 0.04 |
ENST00000359524.5
|
RUNX2
|
runt-related transcription factor 2 |
| chr5_-_68628543 | 0.04 |
ENST00000396496.2
ENST00000511257.1 ENST00000383374.2 |
CCDC125
|
coiled-coil domain containing 125 |
| chr12_+_57853918 | 0.04 |
ENST00000532291.1
ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1
|
GLI family zinc finger 1 |
| chr12_-_8025479 | 0.04 |
ENST00000537557.1
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14 |
| chr19_+_24009879 | 0.04 |
ENST00000354585.4
|
RPSAP58
|
ribosomal protein SA pseudogene 58 |
| chr16_-_79634595 | 0.04 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr11_-_8959758 | 0.04 |
ENST00000531618.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr19_+_46367518 | 0.04 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr1_+_203256898 | 0.04 |
ENST00000433008.1
|
RP11-134P9.3
|
RP11-134P9.3 |
| chr11_+_64052692 | 0.04 |
ENST00000377702.4
|
GPR137
|
G protein-coupled receptor 137 |
| chr9_+_17134980 | 0.04 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr12_+_12202785 | 0.04 |
ENST00000586576.1
ENST00000464885.2 |
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chr12_-_71148357 | 0.04 |
ENST00000378778.1
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R |
| chr11_-_28129656 | 0.04 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chrX_+_77166172 | 0.04 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr7_-_143991230 | 0.04 |
ENST00000543357.1
|
ARHGEF35
|
Rho guanine nucleotide exchange factor (GEF) 35 |
| chr3_+_119217422 | 0.04 |
ENST00000466984.1
|
TIMMDC1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr22_-_16449805 | 0.04 |
ENST00000252835.4
|
OR11H1
|
olfactory receptor, family 11, subfamily H, member 1 |
| chr22_+_40441456 | 0.04 |
ENST00000402203.1
|
TNRC6B
|
trinucleotide repeat containing 6B |
| chrX_+_47077680 | 0.04 |
ENST00000522883.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr6_+_131894284 | 0.04 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr2_-_228579305 | 0.04 |
ENST00000456524.1
|
SLC19A3
|
solute carrier family 19 (thiamine transporter), member 3 |
| chr10_-_65028817 | 0.04 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX21_TBR1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.0 | 0.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.2 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.1 | GO:0043091 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.0 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.0 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.2 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.0 | GO:0034759 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) cellular response to lead ion(GO:0071284) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.0 | GO:1902636 | kinociliary basal body(GO:1902636) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 0.2 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |