Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
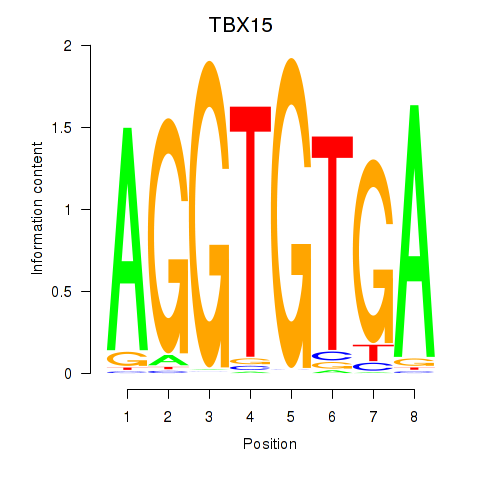
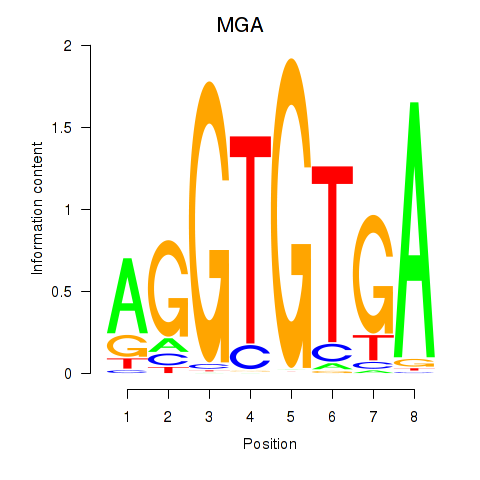
Results for TBX15_MGA
Z-value: 1.49
Transcription factors associated with TBX15_MGA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX15
|
ENSG00000092607.9 | T-box transcription factor 15 |
|
MGA
|
ENSG00000174197.12 | MAX dimerization protein MGA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX15 | hg19_v2_chr1_-_119532127_119532179 | 0.72 | 1.1e-01 | Click! |
| MGA | hg19_v2_chr15_+_41952591_41952672 | -0.30 | 5.7e-01 | Click! |
Activity profile of TBX15_MGA motif
Sorted Z-values of TBX15_MGA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_11907829 | 2.06 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr3_+_167453493 | 2.01 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr11_+_18477369 | 1.96 |
ENST00000396213.3
ENST00000280706.2 |
LDHAL6A
|
lactate dehydrogenase A-like 6A |
| chr2_+_228678550 | 1.65 |
ENST00000409189.3
ENST00000358813.4 |
CCL20
|
chemokine (C-C motif) ligand 20 |
| chr8_-_145060593 | 1.64 |
ENST00000313059.5
ENST00000524918.1 ENST00000313028.7 ENST00000525773.1 |
PARP10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr7_-_120497178 | 1.61 |
ENST00000441017.1
ENST00000424710.1 ENST00000433758.1 |
TSPAN12
|
tetraspanin 12 |
| chr11_-_615570 | 1.53 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr13_-_48575376 | 1.40 |
ENST00000434484.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr19_-_10213335 | 1.21 |
ENST00000592641.1
ENST00000253109.4 |
ANGPTL6
|
angiopoietin-like 6 |
| chr9_+_96717821 | 1.17 |
ENST00000454594.1
|
RP11-231K24.2
|
RP11-231K24.2 |
| chr13_-_48575401 | 1.04 |
ENST00000433022.1
ENST00000544100.1 |
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr14_+_51026844 | 1.02 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr21_+_30502806 | 0.92 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr8_-_99954788 | 0.91 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr3_+_167453026 | 0.90 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr14_+_95078714 | 0.89 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr7_-_81635106 | 0.88 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr2_+_62933001 | 0.86 |
ENST00000263991.5
ENST00000354487.3 |
EHBP1
|
EH domain binding protein 1 |
| chr9_+_139847347 | 0.84 |
ENST00000371632.3
|
LCN12
|
lipocalin 12 |
| chr2_+_62932779 | 0.84 |
ENST00000427809.1
ENST00000405482.1 ENST00000431489.1 |
EHBP1
|
EH domain binding protein 1 |
| chr14_+_51026926 | 0.77 |
ENST00000557735.1
|
ATL1
|
atlastin GTPase 1 |
| chr1_+_155911480 | 0.73 |
ENST00000368318.3
|
RXFP4
|
relaxin/insulin-like family peptide receptor 4 |
| chr5_-_114515734 | 0.73 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chrM_+_10464 | 0.69 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr3_-_120170052 | 0.68 |
ENST00000295633.3
|
FSTL1
|
follistatin-like 1 |
| chr4_+_26578293 | 0.68 |
ENST00000512840.1
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr8_-_99955042 | 0.66 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chr5_-_49737184 | 0.65 |
ENST00000508934.1
ENST00000303221.5 |
EMB
|
embigin |
| chr13_-_48575443 | 0.64 |
ENST00000378654.3
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chrX_+_16804544 | 0.64 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr14_-_45603657 | 0.64 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr3_+_107364769 | 0.64 |
ENST00000449271.1
ENST00000425868.1 ENST00000449213.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr12_+_104324112 | 0.63 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr14_+_75746781 | 0.62 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr2_-_39664405 | 0.62 |
ENST00000341681.5
ENST00000263881.3 |
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr6_-_30815936 | 0.61 |
ENST00000442852.1
|
XXbac-BPG27H4.8
|
XXbac-BPG27H4.8 |
| chr12_-_90024360 | 0.60 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr7_+_107220660 | 0.60 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr19_-_46526304 | 0.60 |
ENST00000008938.4
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr18_-_45457478 | 0.59 |
ENST00000402690.2
ENST00000356825.4 |
SMAD2
|
SMAD family member 2 |
| chr4_+_140222609 | 0.59 |
ENST00000296543.5
ENST00000398947.1 |
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr11_+_107643129 | 0.58 |
ENST00000447610.1
|
AP001024.2
|
Uncharacterized protein |
| chr1_+_172502244 | 0.57 |
ENST00000610051.1
|
SUCO
|
SUN domain containing ossification factor |
| chr11_+_46402482 | 0.56 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr3_+_142315225 | 0.55 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chrM_+_8366 | 0.55 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr11_+_46402297 | 0.55 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr11_-_85430204 | 0.54 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr2_-_74776586 | 0.54 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr10_+_94451574 | 0.53 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr1_+_172502336 | 0.53 |
ENST00000263688.3
|
SUCO
|
SUN domain containing ossification factor |
| chr1_+_150254936 | 0.52 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr12_+_56473939 | 0.52 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr1_+_93544821 | 0.51 |
ENST00000370303.4
|
MTF2
|
metal response element binding transcription factor 2 |
| chr11_+_111749650 | 0.51 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr1_+_26737253 | 0.51 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr12_+_52431016 | 0.51 |
ENST00000553200.1
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr11_-_85779971 | 0.51 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_+_51026743 | 0.51 |
ENST00000358385.6
ENST00000357032.3 ENST00000354525.4 |
ATL1
|
atlastin GTPase 1 |
| chrM_+_10758 | 0.51 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr3_-_167452614 | 0.51 |
ENST00000392750.2
ENST00000464360.1 ENST00000492139.1 ENST00000471885.1 ENST00000470131.1 |
PDCD10
|
programmed cell death 10 |
| chr10_-_16563870 | 0.48 |
ENST00000298943.3
|
C1QL3
|
complement component 1, q subcomponent-like 3 |
| chr14_+_90864504 | 0.48 |
ENST00000544280.2
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr2_-_145090035 | 0.48 |
ENST00000542155.1
ENST00000241391.5 ENST00000463875.2 |
GTDC1
|
glycosyltransferase-like domain containing 1 |
| chr5_+_74807581 | 0.47 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr5_+_74807886 | 0.47 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr1_+_35734616 | 0.47 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr15_-_34502278 | 0.46 |
ENST00000559515.1
ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr2_+_154728426 | 0.46 |
ENST00000392825.3
ENST00000434213.1 |
GALNT13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) |
| chr12_-_111180644 | 0.46 |
ENST00000551676.1
ENST00000550991.1 ENST00000335007.5 ENST00000340766.5 |
PPP1CC
|
protein phosphatase 1, catalytic subunit, gamma isozyme |
| chr1_-_27701307 | 0.45 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr3_+_142315294 | 0.45 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr4_+_183065793 | 0.45 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr9_+_17134980 | 0.45 |
ENST00000380647.3
|
CNTLN
|
centlein, centrosomal protein |
| chr8_-_109260897 | 0.44 |
ENST00000521297.1
ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E
|
eukaryotic translation initiation factor 3, subunit E |
| chr16_-_69418553 | 0.44 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr3_+_107364683 | 0.44 |
ENST00000413213.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr7_+_133261209 | 0.43 |
ENST00000545148.1
|
EXOC4
|
exocyst complex component 4 |
| chr6_+_53794780 | 0.41 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chrX_+_123095546 | 0.41 |
ENST00000371157.3
ENST00000371145.3 ENST00000371144.3 |
STAG2
|
stromal antigen 2 |
| chr11_-_85430088 | 0.41 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr3_-_167452703 | 0.41 |
ENST00000497056.2
ENST00000473645.2 |
PDCD10
|
programmed cell death 10 |
| chr11_+_73358690 | 0.40 |
ENST00000545798.1
ENST00000539157.1 ENST00000546251.1 ENST00000535582.1 ENST00000538227.1 ENST00000543524.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr22_-_41215291 | 0.40 |
ENST00000542412.1
ENST00000544408.1 |
SLC25A17
|
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
| chr17_+_9066252 | 0.39 |
ENST00000436734.1
|
NTN1
|
netrin 1 |
| chr11_+_10772847 | 0.39 |
ENST00000524523.1
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr6_+_108616243 | 0.39 |
ENST00000421954.1
|
LACE1
|
lactation elevated 1 |
| chr5_-_124080203 | 0.39 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr17_-_40833858 | 0.39 |
ENST00000332438.4
|
CCR10
|
chemokine (C-C motif) receptor 10 |
| chr1_-_115124257 | 0.39 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr1_+_93544791 | 0.39 |
ENST00000545708.1
ENST00000540243.1 ENST00000370298.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr4_+_66536248 | 0.39 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr2_+_61108650 | 0.38 |
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr17_-_7120498 | 0.38 |
ENST00000485100.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr13_-_52027134 | 0.38 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr11_+_117947724 | 0.37 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr9_+_131452239 | 0.37 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr11_+_19138670 | 0.36 |
ENST00000446113.2
ENST00000399351.3 |
ZDHHC13
|
zinc finger, DHHC-type containing 13 |
| chr16_-_69418649 | 0.36 |
ENST00000566257.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr12_-_95942563 | 0.36 |
ENST00000549639.1
ENST00000551837.1 |
USP44
|
ubiquitin specific peptidase 44 |
| chr2_+_29033682 | 0.36 |
ENST00000379579.4
ENST00000334056.5 ENST00000449210.1 |
SPDYA
|
speedy/RINGO cell cycle regulator family member A |
| chr9_-_128003606 | 0.34 |
ENST00000324460.6
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr1_+_27113963 | 0.34 |
ENST00000430292.1
|
PIGV
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr3_-_49722523 | 0.34 |
ENST00000448220.1
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr3_+_38206975 | 0.33 |
ENST00000446845.1
ENST00000311806.3 |
OXSR1
|
oxidative stress responsive 1 |
| chr18_-_45457192 | 0.33 |
ENST00000586514.1
ENST00000591214.1 ENST00000589877.1 |
SMAD2
|
SMAD family member 2 |
| chr17_+_30771279 | 0.33 |
ENST00000261712.3
ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr4_+_111397216 | 0.32 |
ENST00000265162.5
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A) |
| chr12_-_122711968 | 0.32 |
ENST00000485724.1
|
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr3_-_167452298 | 0.32 |
ENST00000475915.2
ENST00000462725.2 ENST00000461494.1 |
PDCD10
|
programmed cell death 10 |
| chr3_-_167452262 | 0.32 |
ENST00000487947.2
|
PDCD10
|
programmed cell death 10 |
| chrM_+_8527 | 0.32 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr9_+_17135016 | 0.32 |
ENST00000425824.1
ENST00000262360.5 ENST00000380641.4 |
CNTLN
|
centlein, centrosomal protein |
| chr17_+_57970469 | 0.32 |
ENST00000443572.2
ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| chr11_-_125932685 | 0.32 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr15_+_63481668 | 0.32 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr5_+_68788594 | 0.32 |
ENST00000396442.2
ENST00000380766.2 |
OCLN
|
occludin |
| chr20_+_43343517 | 0.32 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr2_-_61697862 | 0.31 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr21_-_27107198 | 0.31 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr6_-_84937314 | 0.31 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr1_+_199996702 | 0.31 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr15_+_89922345 | 0.30 |
ENST00000558982.1
|
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr6_+_30908747 | 0.30 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr17_+_76311791 | 0.30 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr8_-_17270809 | 0.30 |
ENST00000180173.5
ENST00000521857.1 |
MTMR7
|
myotubularin related protein 7 |
| chr6_-_130536774 | 0.29 |
ENST00000532763.1
|
SAMD3
|
sterile alpha motif domain containing 3 |
| chr20_-_1165319 | 0.29 |
ENST00000429036.1
|
TMEM74B
|
transmembrane protein 74B |
| chr7_-_150675372 | 0.29 |
ENST00000262186.5
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr12_-_76462713 | 0.29 |
ENST00000552056.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr11_+_6260298 | 0.29 |
ENST00000379936.2
|
CNGA4
|
cyclic nucleotide gated channel alpha 4 |
| chr20_-_44516256 | 0.29 |
ENST00000372519.3
|
SPATA25
|
spermatogenesis associated 25 |
| chr12_+_95612006 | 0.29 |
ENST00000551311.1
ENST00000546445.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr1_-_28969517 | 0.29 |
ENST00000263974.4
ENST00000373824.4 |
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr21_-_27107344 | 0.29 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr8_+_103563792 | 0.28 |
ENST00000285402.3
|
ODF1
|
outer dense fiber of sperm tails 1 |
| chr9_-_37384431 | 0.28 |
ENST00000452923.1
|
RP11-397D12.4
|
RP11-397D12.4 |
| chr1_-_113257905 | 0.28 |
ENST00000464951.1
|
PPM1J
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr13_+_76378357 | 0.28 |
ENST00000489941.2
ENST00000525373.1 |
LMO7
|
LIM domain 7 |
| chr7_+_141438393 | 0.28 |
ENST00000484178.1
ENST00000473783.1 ENST00000481508.1 |
SSBP1
|
single-stranded DNA binding protein 1, mitochondrial |
| chr9_-_75567962 | 0.27 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr2_+_43864387 | 0.27 |
ENST00000282406.4
|
PLEKHH2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr1_+_26737292 | 0.27 |
ENST00000254231.4
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr9_+_75766652 | 0.27 |
ENST00000257497.6
|
ANXA1
|
annexin A1 |
| chr13_+_76378305 | 0.27 |
ENST00000526371.1
ENST00000526528.1 |
LMO7
|
LIM domain 7 |
| chr12_-_123560608 | 0.27 |
ENST00000541244.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr10_+_98592674 | 0.27 |
ENST00000356016.3
ENST00000371097.4 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chr19_-_44405941 | 0.27 |
ENST00000587128.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr20_+_44657845 | 0.27 |
ENST00000243964.3
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5 |
| chr15_-_33180439 | 0.27 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr2_-_160472952 | 0.27 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr14_-_55658323 | 0.27 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr19_+_46002868 | 0.27 |
ENST00000396735.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr12_+_58148842 | 0.27 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr2_+_169312725 | 0.26 |
ENST00000392687.4
|
CERS6
|
ceramide synthase 6 |
| chr17_-_7120525 | 0.26 |
ENST00000447163.1
ENST00000399506.2 ENST00000302955.6 |
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr2_+_102758751 | 0.26 |
ENST00000442590.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr1_-_113258090 | 0.26 |
ENST00000309276.6
|
PPM1J
|
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr21_-_46962379 | 0.25 |
ENST00000311124.4
ENST00000380010.4 |
SLC19A1
|
solute carrier family 19 (folate transporter), member 1 |
| chr17_+_57784997 | 0.25 |
ENST00000537567.1
ENST00000539763.1 ENST00000587945.1 ENST00000536180.1 ENST00000589823.2 ENST00000592106.1 ENST00000591315.1 ENST00000545362.1 |
VMP1
|
vacuole membrane protein 1 |
| chr20_+_2517253 | 0.25 |
ENST00000358864.1
|
TMC2
|
transmembrane channel-like 2 |
| chr14_-_98444386 | 0.25 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr2_-_37544209 | 0.25 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr10_+_95256356 | 0.25 |
ENST00000371485.3
|
CEP55
|
centrosomal protein 55kDa |
| chr19_-_45748628 | 0.25 |
ENST00000590022.1
|
AC006126.4
|
AC006126.4 |
| chr5_+_154320623 | 0.24 |
ENST00000523037.1
ENST00000265229.8 ENST00000439747.3 ENST00000522038.1 |
MRPL22
|
mitochondrial ribosomal protein L22 |
| chr1_-_25256368 | 0.24 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr3_+_160117418 | 0.24 |
ENST00000465903.1
ENST00000485645.1 ENST00000360111.2 ENST00000472991.1 ENST00000467468.1 ENST00000469762.1 ENST00000489573.1 ENST00000462787.1 ENST00000490207.1 ENST00000485867.1 |
SMC4
|
structural maintenance of chromosomes 4 |
| chrX_-_99891796 | 0.24 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr2_-_160473114 | 0.24 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr12_-_13529594 | 0.24 |
ENST00000539026.1
|
C12orf36
|
chromosome 12 open reading frame 36 |
| chr1_+_199996733 | 0.24 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chrM_+_9207 | 0.24 |
ENST00000362079.2
|
MT-CO3
|
mitochondrially encoded cytochrome c oxidase III |
| chr9_-_73029540 | 0.24 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr14_-_55658252 | 0.24 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr1_+_8378140 | 0.23 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr10_-_105238997 | 0.23 |
ENST00000369783.4
|
CALHM3
|
calcium homeostasis modulator 3 |
| chr14_+_45553296 | 0.23 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr17_-_39203519 | 0.23 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr11_-_85430163 | 0.23 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr11_+_73358594 | 0.23 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chrX_+_123094672 | 0.23 |
ENST00000354548.5
ENST00000458700.1 |
STAG2
|
stromal antigen 2 |
| chr8_+_22019168 | 0.23 |
ENST00000318561.3
ENST00000521315.1 ENST00000437090.2 ENST00000520605.1 ENST00000522109.1 ENST00000524255.1 ENST00000523296.1 ENST00000518615.1 |
SFTPC
|
surfactant protein C |
| chr19_-_51054299 | 0.23 |
ENST00000599957.1
|
LRRC4B
|
leucine rich repeat containing 4B |
| chr7_-_6523688 | 0.23 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr10_-_74856608 | 0.23 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chrX_+_123095155 | 0.23 |
ENST00000371160.1
ENST00000435103.1 |
STAG2
|
stromal antigen 2 |
| chrX_+_21958814 | 0.23 |
ENST00000379404.1
ENST00000415881.2 |
SMS
|
spermine synthase |
| chr2_+_99953816 | 0.22 |
ENST00000289371.6
|
EIF5B
|
eukaryotic translation initiation factor 5B |
| chr5_+_56469843 | 0.22 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr9_+_131451480 | 0.22 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr10_+_111765562 | 0.22 |
ENST00000360162.3
|
ADD3
|
adducin 3 (gamma) |
| chr11_+_111749942 | 0.22 |
ENST00000260276.3
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr9_-_74979420 | 0.22 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr8_-_124253576 | 0.22 |
ENST00000276704.4
|
C8orf76
|
chromosome 8 open reading frame 76 |
| chr21_-_16374688 | 0.22 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr2_+_172864490 | 0.22 |
ENST00000315796.4
|
METAP1D
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr7_-_99527243 | 0.22 |
ENST00000312891.2
|
GJC3
|
gap junction protein, gamma 3, 30.2kDa |
| chr2_+_61108771 | 0.22 |
ENST00000394479.3
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr7_+_27282319 | 0.22 |
ENST00000222761.3
|
EVX1
|
even-skipped homeobox 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX15_MGA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.5 | 1.6 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.5 | 1.5 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.4 | 2.1 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.3 | 1.6 | GO:0010847 | regulation of chromatin assembly(GO:0010847) protein auto-ADP-ribosylation(GO:0070213) |
| 0.3 | 1.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.3 | 1.6 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.2 | 0.6 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.2 | 0.8 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.2 | 0.6 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) |
| 0.2 | 1.1 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 0.9 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.2 | 0.5 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.2 | 1.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.6 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.4 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.4 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.1 | 0.9 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.2 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.1 | 0.3 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.9 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 0.6 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 0.5 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.1 | 0.3 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.4 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.1 | 0.6 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 1.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.8 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.5 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.1 | 0.7 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.5 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.5 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.6 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 | 0.6 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.2 | GO:2000987 | cell communication by chemical coupling(GO:0010643) positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.9 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.3 | GO:0098838 | reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.2 | GO:0043132 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) NAD transport(GO:0043132) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 0.5 | GO:0071287 | cellular response to manganese ion(GO:0071287) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.3 | GO:0044254 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.1 | 0.2 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.1 | 0.4 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 1.7 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 1.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.4 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.2 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:1905069 | allantois development(GO:1905069) |
| 0.0 | 0.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.3 | GO:0072093 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.0 | 1.1 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.2 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.4 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.0 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 3.5 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 0.9 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.3 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.0 | GO:0035284 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.3 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.6 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.3 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.0 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.5 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.6 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.2 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.1 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.2 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 1.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0061087 | regulation of histone H3-K27 methylation(GO:0061085) positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.4 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.6 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 0.9 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.5 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.4 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.6 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.5 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.2 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.1 | 2.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 1.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 0.0 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.0 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) STAGA complex(GO:0030914) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.1 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.5 | 1.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.3 | 0.9 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.2 | 2.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.5 | GO:0004584 | dolichyl-phosphate-mannose-glycolipid alpha-mannosyltransferase activity(GO:0004584) |
| 0.2 | 0.6 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.7 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.4 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.1 | 0.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 2.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.4 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.3 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.2 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.8 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.8 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.2 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.9 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 1.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.9 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.0 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 1.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.6 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.1 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.0 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.5 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 3.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 2.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 2.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 1.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |