Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
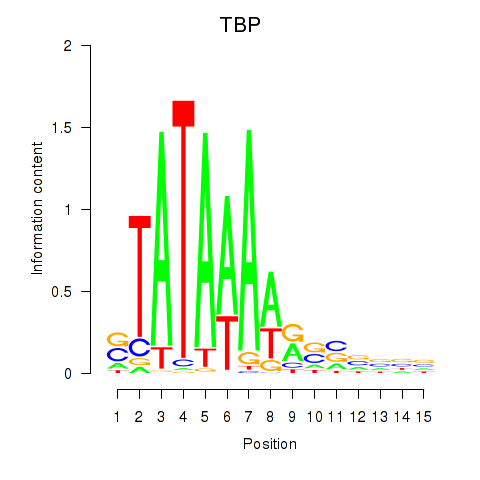
Results for TBP
Z-value: 2.69
Transcription factors associated with TBP
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBP
|
ENSG00000112592.8 | TATA-box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBP | hg19_v2_chr6_+_170863353_170863390 | 0.72 | 1.1e-01 | Click! |
Activity profile of TBP motif
Sorted Z-values of TBP motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_615570 | 3.36 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr10_+_91152303 | 3.30 |
ENST00000371804.3
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr6_-_26189304 | 2.36 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr9_-_35658007 | 2.17 |
ENST00000602361.1
|
RMRP
|
RNA component of mitochondrial RNA processing endoribonuclease |
| chr6_+_26104104 | 2.14 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr16_+_2014993 | 2.11 |
ENST00000564014.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr6_-_26043885 | 2.05 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr10_+_91087651 | 1.68 |
ENST00000371818.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr12_-_121476959 | 1.61 |
ENST00000339275.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr6_+_26156551 | 1.60 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr4_-_169239921 | 1.57 |
ENST00000514995.1
ENST00000393743.3 |
DDX60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr11_+_18287721 | 1.49 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr12_-_121477039 | 1.46 |
ENST00000257570.5
|
OASL
|
2'-5'-oligoadenylate synthetase-like |
| chr6_+_27107053 | 1.38 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr6_+_26045603 | 1.36 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr17_-_39023462 | 1.30 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr6_+_27114861 | 1.28 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr6_-_26235206 | 1.25 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr1_-_149785236 | 1.22 |
ENST00000331491.1
|
HIST2H3D
|
histone cluster 2, H3d |
| chr1_+_33546714 | 1.17 |
ENST00000294517.6
ENST00000358680.3 ENST00000373443.3 ENST00000398167.1 |
ADC
|
arginine decarboxylase |
| chr17_-_34207295 | 1.15 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr6_+_26217159 | 1.15 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr16_-_67965756 | 1.14 |
ENST00000571044.1
ENST00000571605.1 |
CTRL
|
chymotrypsin-like |
| chr12_-_118796971 | 1.10 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr11_-_1619524 | 1.10 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr11_+_18287801 | 1.07 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr19_-_6720686 | 1.04 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr1_+_153330322 | 1.00 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr7_-_92777606 | 0.99 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr4_-_74904398 | 0.98 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr18_+_68002675 | 0.93 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr3_-_170588163 | 0.89 |
ENST00000295830.8
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr17_+_39411636 | 0.88 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr6_-_27840099 | 0.86 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr16_+_2014941 | 0.85 |
ENST00000531523.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr17_+_80186908 | 0.80 |
ENST00000582743.1
ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr9_+_92219919 | 0.80 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr14_-_50506589 | 0.80 |
ENST00000553914.2
|
RP11-58E21.3
|
RP11-58E21.3 |
| chr1_-_220101944 | 0.80 |
ENST00000366926.3
ENST00000536992.1 |
SLC30A10
|
solute carrier family 30, member 10 |
| chr7_-_75401513 | 0.77 |
ENST00000005180.4
|
CCL26
|
chemokine (C-C motif) ligand 26 |
| chr4_+_74702214 | 0.77 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr12_+_52695617 | 0.76 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr1_-_228613026 | 0.76 |
ENST00000366696.1
|
HIST3H3
|
histone cluster 3, H3 |
| chr17_+_39421591 | 0.75 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr9_+_130911723 | 0.73 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr4_+_74735102 | 0.70 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chrX_-_133792480 | 0.69 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr4_-_74853897 | 0.68 |
ENST00000296028.3
|
PPBP
|
pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) |
| chr6_+_159290917 | 0.67 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr19_+_10381769 | 0.67 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr6_-_27841289 | 0.66 |
ENST00000355981.2
|
HIST1H4L
|
histone cluster 1, H4l |
| chr3_-_170587815 | 0.66 |
ENST00000466674.1
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr9_+_130911770 | 0.66 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr19_+_36347787 | 0.64 |
ENST00000347900.6
ENST00000360202.5 |
KIRREL2
|
kin of IRRE like 2 (Drosophila) |
| chr17_+_74729060 | 0.61 |
ENST00000587459.1
|
RP11-318A15.7
|
Uncharacterized protein |
| chr1_-_89488510 | 0.60 |
ENST00000564665.1
ENST00000370481.4 |
GBP3
|
guanylate binding protein 3 |
| chr4_+_74606223 | 0.59 |
ENST00000307407.3
ENST00000401931.1 |
IL8
|
interleukin 8 |
| chr1_-_16763911 | 0.58 |
ENST00000375577.1
ENST00000335496.1 |
SPATA21
|
spermatogenesis associated 21 |
| chr18_-_61329118 | 0.57 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr22_+_19706958 | 0.57 |
ENST00000395109.2
|
SEPT5
|
septin 5 |
| chr17_-_38928414 | 0.56 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr3_+_57882061 | 0.56 |
ENST00000461354.1
ENST00000466255.1 |
SLMAP
|
sarcolemma associated protein |
| chr19_-_53636125 | 0.56 |
ENST00000601493.1
ENST00000599261.1 ENST00000597503.1 ENST00000500065.4 ENST00000243643.4 ENST00000594011.1 ENST00000455735.2 ENST00000595193.1 ENST00000448501.1 ENST00000421033.1 ENST00000440291.1 ENST00000595813.1 ENST00000600574.1 ENST00000596051.1 ENST00000601110.1 |
ZNF415
|
zinc finger protein 415 |
| chr3_+_149192475 | 0.55 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr11_+_45168182 | 0.55 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chr7_+_6121296 | 0.54 |
ENST00000428901.1
|
AC004895.4
|
AC004895.4 |
| chr16_+_57406368 | 0.54 |
ENST00000006053.6
ENST00000563383.1 |
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr17_+_76183398 | 0.54 |
ENST00000409257.5
|
AFMID
|
arylformamidase |
| chr11_-_102651343 | 0.54 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr14_+_56127960 | 0.52 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr3_-_170587974 | 0.52 |
ENST00000463836.1
|
RPL22L1
|
ribosomal protein L22-like 1 |
| chr6_-_26197478 | 0.51 |
ENST00000356476.2
|
HIST1H3D
|
histone cluster 1, H3d |
| chr12_+_11905413 | 0.51 |
ENST00000545027.1
|
ETV6
|
ets variant 6 |
| chr10_+_54074033 | 0.51 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr6_+_27833034 | 0.50 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr15_-_41166414 | 0.49 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr2_+_33661382 | 0.48 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr14_+_56127989 | 0.47 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr17_+_76183432 | 0.47 |
ENST00000591256.1
ENST00000589256.1 ENST00000588800.1 ENST00000591952.1 ENST00000327898.5 ENST00000586542.1 ENST00000586731.1 |
AFMID
|
arylformamidase |
| chr6_-_76203345 | 0.47 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr20_+_30193083 | 0.46 |
ENST00000376112.3
ENST00000376105.3 |
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein |
| chr4_+_108815402 | 0.46 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr10_+_48247669 | 0.45 |
ENST00000457620.1
|
FAM25G
|
family with sequence similarity 25, member G |
| chr1_-_156675564 | 0.45 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr1_+_37940153 | 0.45 |
ENST00000373087.6
|
ZC3H12A
|
zinc finger CCCH-type containing 12A |
| chr10_-_47181681 | 0.45 |
ENST00000452267.1
|
FAM25B
|
family with sequence similarity 25, member B |
| chr18_-_53735601 | 0.45 |
ENST00000589754.1
|
CTD-2008L17.2
|
CTD-2008L17.2 |
| chr6_-_26032288 | 0.44 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr7_+_75024337 | 0.44 |
ENST00000450434.1
|
TRIM73
|
tripartite motif containing 73 |
| chr1_+_43424698 | 0.43 |
ENST00000431759.1
|
SLC2A1-AS1
|
SLC2A1 antisense RNA 1 |
| chr2_+_39893043 | 0.43 |
ENST00000281961.2
|
TMEM178A
|
transmembrane protein 178A |
| chr1_+_198189921 | 0.42 |
ENST00000391974.3
|
NEK7
|
NIMA-related kinase 7 |
| chr19_+_496454 | 0.42 |
ENST00000346144.4
ENST00000215637.3 ENST00000382683.4 |
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1 |
| chr9_+_138413277 | 0.42 |
ENST00000263598.2
ENST00000371781.3 |
LCN1
|
lipocalin 1 |
| chr8_+_145321517 | 0.41 |
ENST00000340210.1
|
SCXB
|
scleraxis homolog B (mouse) |
| chr19_+_46367518 | 0.41 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr12_-_118796910 | 0.40 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_27861190 | 0.40 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr6_-_52859968 | 0.39 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr6_-_78173490 | 0.39 |
ENST00000369947.2
|
HTR1B
|
5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled |
| chr17_-_39341594 | 0.39 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr15_+_45722727 | 0.39 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr1_+_92414952 | 0.39 |
ENST00000449584.1
ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT
|
bromodomain, testis-specific |
| chr4_-_140098339 | 0.39 |
ENST00000394235.2
|
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr7_+_107301447 | 0.38 |
ENST00000440056.1
|
SLC26A4
|
solute carrier family 26 (anion exchanger), member 4 |
| chr16_+_2880369 | 0.38 |
ENST00000572863.1
|
ZG16B
|
zymogen granule protein 16B |
| chr11_+_64937625 | 0.38 |
ENST00000377185.2
|
SPDYC
|
speedy/RINGO cell cycle regulator family member C |
| chr5_-_57756087 | 0.37 |
ENST00000274289.3
|
PLK2
|
polo-like kinase 2 |
| chr5_-_34043310 | 0.37 |
ENST00000231338.7
|
C1QTNF3
|
C1q and tumor necrosis factor related protein 3 |
| chr19_+_4769117 | 0.37 |
ENST00000540211.1
ENST00000317292.3 ENST00000586721.1 ENST00000592709.1 ENST00000588711.1 ENST00000589639.1 ENST00000591008.1 ENST00000592663.1 ENST00000588758.1 |
MIR7-3HG
|
MIR7-3 host gene (non-protein coding) |
| chr18_+_21718924 | 0.37 |
ENST00000399496.3
|
CABYR
|
calcium binding tyrosine-(Y)-phosphorylation regulated |
| chr3_-_131756559 | 0.37 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr3_+_101546827 | 0.37 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr12_-_11184006 | 0.37 |
ENST00000390675.2
|
TAS2R31
|
taste receptor, type 2, member 31 |
| chr17_-_73150629 | 0.37 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr10_+_7745232 | 0.36 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr19_-_8386238 | 0.36 |
ENST00000301457.2
|
NDUFA7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa |
| chr10_-_74856608 | 0.36 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr1_-_11907829 | 0.36 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr6_+_159291090 | 0.36 |
ENST00000367073.4
ENST00000608817.1 |
C6orf99
|
chromosome 6 open reading frame 99 |
| chr7_-_7575477 | 0.35 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr14_+_65007177 | 0.35 |
ENST00000247207.6
|
HSPA2
|
heat shock 70kDa protein 2 |
| chr19_+_840963 | 0.35 |
ENST00000234347.5
|
PRTN3
|
proteinase 3 |
| chrX_-_119077695 | 0.34 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr19_+_13049413 | 0.34 |
ENST00000316448.5
ENST00000588454.1 |
CALR
|
calreticulin |
| chr1_-_179457805 | 0.34 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chrX_+_18725758 | 0.33 |
ENST00000472826.1
ENST00000544635.1 ENST00000496075.2 |
PPEF1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr5_-_79946820 | 0.33 |
ENST00000604882.1
|
MTRNR2L2
|
MT-RNR2-like 2 |
| chr8_+_86089460 | 0.33 |
ENST00000418930.2
|
E2F5
|
E2F transcription factor 5, p130-binding |
| chr22_-_40929812 | 0.32 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr6_+_64281906 | 0.32 |
ENST00000370651.3
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr6_-_26018007 | 0.32 |
ENST00000244573.3
|
HIST1H1A
|
histone cluster 1, H1a |
| chr7_+_108210012 | 0.32 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr6_-_26199499 | 0.32 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr19_-_33793430 | 0.32 |
ENST00000498907.2
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr11_-_102714534 | 0.32 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr12_-_21757774 | 0.30 |
ENST00000261195.2
|
GYS2
|
glycogen synthase 2 (liver) |
| chr19_+_46732988 | 0.29 |
ENST00000437936.1
|
IGFL1
|
IGF-like family member 1 |
| chr5_-_153857819 | 0.29 |
ENST00000231121.2
|
HAND1
|
heart and neural crest derivatives expressed 1 |
| chr2_-_224467093 | 0.29 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr1_-_8086343 | 0.29 |
ENST00000474874.1
ENST00000469499.1 ENST00000377482.5 |
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr3_-_96337000 | 0.29 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr10_-_94301107 | 0.29 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chrX_-_119445306 | 0.28 |
ENST00000371369.4
ENST00000440464.1 ENST00000519908.1 |
TMEM255A
|
transmembrane protein 255A |
| chr6_-_26199471 | 0.27 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr11_-_62432641 | 0.27 |
ENST00000528405.1
ENST00000524958.1 ENST00000525675.1 |
RP11-831H9.11
C11orf48
|
Uncharacterized protein chromosome 11 open reading frame 48 |
| chr1_-_116926718 | 0.27 |
ENST00000598661.1
|
AL136376.1
|
Uncharacterized protein |
| chr7_+_29186174 | 0.26 |
ENST00000439384.1
|
CHN2
|
chimerin 2 |
| chr9_-_33447584 | 0.26 |
ENST00000297991.4
|
AQP3
|
aquaporin 3 (Gill blood group) |
| chr1_-_42630389 | 0.26 |
ENST00000357001.2
|
GUCA2A
|
guanylate cyclase activator 2A (guanylin) |
| chr10_+_112327425 | 0.26 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr7_-_55640141 | 0.25 |
ENST00000452832.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chrX_+_41548220 | 0.25 |
ENST00000378142.4
|
GPR34
|
G protein-coupled receptor 34 |
| chr10_+_6130946 | 0.25 |
ENST00000379888.4
|
RBM17
|
RNA binding motif protein 17 |
| chr11_-_9025541 | 0.25 |
ENST00000525100.1
ENST00000309166.3 ENST00000531090.1 |
NRIP3
|
nuclear receptor interacting protein 3 |
| chr6_+_136172820 | 0.25 |
ENST00000308191.6
|
PDE7B
|
phosphodiesterase 7B |
| chr7_+_133615169 | 0.24 |
ENST00000541309.1
|
EXOC4
|
exocyst complex component 4 |
| chr13_+_43597269 | 0.24 |
ENST00000379221.2
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr21_-_43735446 | 0.24 |
ENST00000398431.2
|
TFF3
|
trefoil factor 3 (intestinal) |
| chr19_+_12944722 | 0.24 |
ENST00000591495.1
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr1_+_17575584 | 0.24 |
ENST00000375460.3
|
PADI3
|
peptidyl arginine deiminase, type III |
| chr8_+_1993152 | 0.24 |
ENST00000262113.4
|
MYOM2
|
myomesin 2 |
| chr6_+_64282447 | 0.24 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr21_+_33031935 | 0.24 |
ENST00000270142.6
ENST00000389995.4 |
SOD1
|
superoxide dismutase 1, soluble |
| chr22_+_19466980 | 0.23 |
ENST00000407835.1
ENST00000438587.1 |
CDC45
|
cell division cycle 45 |
| chr2_+_85132749 | 0.23 |
ENST00000233143.4
|
TMSB10
|
thymosin beta 10 |
| chr1_-_169703203 | 0.23 |
ENST00000333360.7
ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE
|
selectin E |
| chr3_-_165555200 | 0.23 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr2_-_99917639 | 0.22 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr3_+_186285192 | 0.22 |
ENST00000439351.1
|
DNAJB11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr3_+_70048881 | 0.22 |
ENST00000483525.1
|
RP11-460N16.1
|
RP11-460N16.1 |
| chr6_+_27806319 | 0.22 |
ENST00000606613.1
ENST00000396980.3 |
HIST1H2BN
|
histone cluster 1, H2bn |
| chr16_+_69796209 | 0.22 |
ENST00000359154.2
ENST00000561780.1 ENST00000563659.1 ENST00000448661.1 |
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr6_+_64346386 | 0.22 |
ENST00000509330.1
|
PHF3
|
PHD finger protein 3 |
| chr6_+_26204825 | 0.22 |
ENST00000360441.4
|
HIST1H4E
|
histone cluster 1, H4e |
| chr1_+_86046433 | 0.22 |
ENST00000451137.2
|
CYR61
|
cysteine-rich, angiogenic inducer, 61 |
| chr17_-_33700626 | 0.21 |
ENST00000308377.4
ENST00000394566.1 ENST00000589811.1 ENST00000430814.1 ENST00000427966.1 ENST00000588579.1 ENST00000591682.1 ENST00000441608.2 ENST00000592122.1 |
SLFN11
|
schlafen family member 11 |
| chr19_-_53238260 | 0.21 |
ENST00000453741.2
ENST00000602162.1 ENST00000601643.1 ENST00000596702.1 ENST00000600943.1 ENST00000543227.1 ENST00000540744.1 |
ZNF611
|
zinc finger protein 611 |
| chr4_+_169842707 | 0.21 |
ENST00000503290.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr22_+_25202232 | 0.21 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr1_+_202830876 | 0.21 |
ENST00000456105.2
|
RP11-480I12.7
|
RP11-480I12.7 |
| chr17_-_79849438 | 0.20 |
ENST00000331204.4
ENST00000505490.2 |
ALYREF
|
Aly/REF export factor |
| chr2_-_182545603 | 0.20 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr20_-_43883197 | 0.20 |
ENST00000338380.2
|
SLPI
|
secretory leukocyte peptidase inhibitor |
| chr12_+_58013693 | 0.20 |
ENST00000320442.4
ENST00000379218.2 |
SLC26A10
|
solute carrier family 26, member 10 |
| chr7_-_134143841 | 0.20 |
ENST00000285930.4
|
AKR1B1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr21_-_43786634 | 0.19 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr17_-_38956205 | 0.19 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chrX_-_48814810 | 0.19 |
ENST00000376488.3
ENST00000396743.3 ENST00000156084.4 |
OTUD5
|
OTU domain containing 5 |
| chr22_+_39916558 | 0.19 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr1_+_46049706 | 0.19 |
ENST00000527470.1
ENST00000525515.1 ENST00000537798.1 ENST00000402363.3 ENST00000528238.1 ENST00000350030.3 ENST00000470768.1 ENST00000372052.4 ENST00000351223.3 |
NASP
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr22_+_19467261 | 0.19 |
ENST00000455750.1
ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45
|
cell division cycle 45 |
| chr5_-_54988559 | 0.19 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr6_-_3227877 | 0.19 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr18_+_9102669 | 0.19 |
ENST00000497577.2
|
NDUFV2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr19_+_8386371 | 0.19 |
ENST00000600659.2
|
RPS28
|
ribosomal protein S28 |
| chr8_+_1993173 | 0.19 |
ENST00000523438.1
|
MYOM2
|
myomesin 2 |
| chr11_+_46958248 | 0.19 |
ENST00000536126.1
ENST00000278460.7 ENST00000378618.2 ENST00000395460.2 ENST00000378615.3 ENST00000543718.1 |
C11orf49
|
chromosome 11 open reading frame 49 |
| chr4_+_119199864 | 0.18 |
ENST00000602414.1
ENST00000602520.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr13_+_27844464 | 0.18 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of TBP
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.8 | 3.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.4 | 1.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.3 | 1.0 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.3 | 1.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 | 2.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.3 | 1.4 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.3 | 1.6 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 0.5 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.2 | 0.5 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 0.2 | 0.5 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.2 | 0.5 | GO:2000627 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.1 | 0.6 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.4 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 1.7 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 3.1 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 0.8 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.4 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.1 | 0.7 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.8 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.3 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.3 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.4 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.1 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 2.6 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 0.3 | GO:2000508 | regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.1 | 0.2 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 0.8 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 1.0 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.2 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.1 | 0.9 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.6 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 0.8 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.2 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0060467 | egg activation(GO:0007343) negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.1 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.4 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.0 | 0.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 1.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.2 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.3 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.1 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.2 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0045083 | negative regulation of interleukin-12 biosynthetic process(GO:0045083) |
| 0.0 | 0.2 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 3.3 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.0 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.0 | 0.1 | GO:0018277 | protein deamination(GO:0018277) |
| 0.0 | 0.2 | GO:0045943 | regulation of transcription from RNA polymerase I promoter(GO:0006356) positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.3 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 4.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:0034694 | response to prostaglandin(GO:0034694) |
| 0.0 | 0.7 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.3 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.5 | GO:0061000 | negative regulation of dendritic spine development(GO:0061000) |
| 0.0 | 0.3 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.3 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) lung vasculature development(GO:0060426) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.1 | GO:1904796 | regulation of core promoter binding(GO:1904796) positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.2 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.3 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0060323 | head morphogenesis(GO:0060323) face morphogenesis(GO:0060325) |
| 0.0 | 2.2 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.5 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.5 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.5 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.0 | GO:0042322 | positive regulation of growth rate(GO:0040010) negative regulation of circadian sleep/wake cycle, REM sleep(GO:0042322) regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.4 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0045907 | positive regulation of vasoconstriction(GO:0045907) |
| 0.0 | 0.2 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.4 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 2.6 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 3.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 3.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 3.7 | GO:0018995 | host(GO:0018995) host cell(GO:0043657) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 2.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.5 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 2.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.0 | GO:0070702 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.3 | 3.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.2 | 3.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 0.8 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.2 | 0.9 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.2 | 1.0 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.1 | 0.3 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.1 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.8 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.5 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 3.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.4 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.2 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.4 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.2 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.4 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 1.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0047017 | geranylgeranyl reductase activity(GO:0045550) prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.2 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.4 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.5 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 2.9 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 2.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.4 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.5 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.0 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.0 | GO:0016608 | growth hormone-releasing hormone activity(GO:0016608) ghrelin receptor binding(GO:0031768) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 1.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.0 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 2.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 3.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.2 | 11.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 3.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 4.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 8.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |