Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
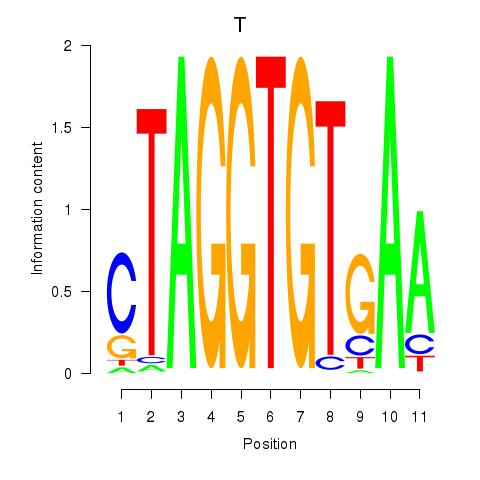
Results for T
Z-value: 0.56
Transcription factors associated with T
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
T
|
ENSG00000164458.5 | T |
Activity profile of T motif
Sorted Z-values of T motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_81635106 | 0.42 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr17_+_34431212 | 0.30 |
ENST00000394495.1
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr6_+_24126350 | 0.28 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr19_+_10197463 | 0.26 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr3_-_52488048 | 0.25 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr2_+_102758751 | 0.24 |
ENST00000442590.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr8_-_99955042 | 0.20 |
ENST00000519420.1
|
STK3
|
serine/threonine kinase 3 |
| chrX_+_69501943 | 0.20 |
ENST00000509895.1
ENST00000374473.2 ENST00000276066.4 |
RAB41
|
RAB41, member RAS oncogene family |
| chr15_-_33180439 | 0.20 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr8_-_99954788 | 0.20 |
ENST00000523601.1
|
STK3
|
serine/threonine kinase 3 |
| chr1_-_204165610 | 0.20 |
ENST00000367194.4
|
KISS1
|
KiSS-1 metastasis-suppressor |
| chr3_-_114035026 | 0.19 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr7_+_107220660 | 0.19 |
ENST00000465919.1
ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29
|
B-cell receptor-associated protein 29 |
| chr2_-_152118352 | 0.16 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr17_+_34430980 | 0.16 |
ENST00000250151.4
|
CCL4
|
chemokine (C-C motif) ligand 4 |
| chr2_+_102758271 | 0.16 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr3_+_38307293 | 0.15 |
ENST00000311856.4
|
SLC22A13
|
solute carrier family 22 (organic anion/urate transporter), member 13 |
| chr12_-_52845910 | 0.15 |
ENST00000252252.3
|
KRT6B
|
keratin 6B |
| chr17_+_7533439 | 0.15 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr9_+_40028620 | 0.14 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr17_-_46657473 | 0.14 |
ENST00000332503.5
|
HOXB4
|
homeobox B4 |
| chr3_+_167453493 | 0.14 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr16_+_72459838 | 0.14 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr2_+_176972000 | 0.14 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chr14_+_75746781 | 0.14 |
ENST00000555347.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr2_+_102758210 | 0.13 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr1_-_25256368 | 0.13 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr20_-_44298878 | 0.13 |
ENST00000324384.3
ENST00000356562.2 |
WFDC11
|
WAP four-disulfide core domain 11 |
| chr19_+_3721719 | 0.12 |
ENST00000589378.1
ENST00000382008.3 |
TJP3
|
tight junction protein 3 |
| chr12_+_8849773 | 0.12 |
ENST00000541044.1
|
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr7_-_994302 | 0.12 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr2_+_234601512 | 0.12 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr2_+_171571827 | 0.12 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr2_+_9778872 | 0.12 |
ENST00000478468.1
|
RP11-521D12.1
|
RP11-521D12.1 |
| chr6_+_159290917 | 0.12 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr2_+_58134756 | 0.12 |
ENST00000435505.2
ENST00000417641.2 |
VRK2
|
vaccinia related kinase 2 |
| chr14_+_85994943 | 0.11 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr19_-_6690723 | 0.11 |
ENST00000601008.1
|
C3
|
complement component 3 |
| chr12_+_8850277 | 0.11 |
ENST00000539923.1
ENST00000537189.1 |
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr12_+_14572070 | 0.11 |
ENST00000545769.1
ENST00000428217.2 ENST00000396279.2 ENST00000542514.1 ENST00000536279.1 |
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr11_+_61197508 | 0.10 |
ENST00000541135.1
ENST00000301761.2 |
RP11-286N22.8
SDHAF2
|
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chr16_-_88717423 | 0.10 |
ENST00000568278.1
ENST00000569359.1 ENST00000567174.1 |
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr14_-_45603657 | 0.10 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr3_-_52002194 | 0.10 |
ENST00000466412.1
|
PCBP4
|
poly(rC) binding protein 4 |
| chr17_-_19281203 | 0.10 |
ENST00000487415.2
|
B9D1
|
B9 protein domain 1 |
| chr11_+_111412271 | 0.10 |
ENST00000528102.1
|
LAYN
|
layilin |
| chr20_-_1309809 | 0.10 |
ENST00000360779.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr2_+_33661382 | 0.10 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr11_-_8739566 | 0.10 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr6_-_106773291 | 0.10 |
ENST00000343245.3
|
ATG5
|
autophagy related 5 |
| chr15_+_78832747 | 0.09 |
ENST00000560217.1
ENST00000044462.7 ENST00000559082.1 ENST00000559948.1 ENST00000413382.2 ENST00000559146.1 ENST00000558281.1 |
PSMA4
|
proteasome (prosome, macropain) subunit, alpha type, 4 |
| chr3_-_133614467 | 0.09 |
ENST00000469959.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr4_+_26322409 | 0.09 |
ENST00000514807.1
ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chrX_+_16804544 | 0.09 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr9_+_131452239 | 0.09 |
ENST00000372688.4
ENST00000372686.5 |
SET
|
SET nuclear oncogene |
| chr1_-_115124257 | 0.09 |
ENST00000369541.3
|
BCAS2
|
breast carcinoma amplified sequence 2 |
| chr4_-_140005443 | 0.09 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chrX_+_133594168 | 0.09 |
ENST00000298556.7
|
HPRT1
|
hypoxanthine phosphoribosyltransferase 1 |
| chr5_+_158654712 | 0.08 |
ENST00000520323.1
|
CTB-11I22.2
|
CTB-11I22.2 |
| chr21_-_27107198 | 0.08 |
ENST00000400094.1
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr5_+_67576062 | 0.08 |
ENST00000523807.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr1_+_35225339 | 0.07 |
ENST00000339480.1
|
GJB4
|
gap junction protein, beta 4, 30.3kDa |
| chrX_-_106146547 | 0.07 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr21_-_45078019 | 0.07 |
ENST00000542962.1
|
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr11_+_61197572 | 0.07 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr15_-_81195510 | 0.07 |
ENST00000561295.1
|
RP11-351M8.1
|
Uncharacterized protein |
| chr14_+_95078714 | 0.07 |
ENST00000393078.3
ENST00000393080.4 ENST00000467132.1 |
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr11_-_85779971 | 0.06 |
ENST00000393346.3
|
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr5_+_50678921 | 0.06 |
ENST00000230658.7
|
ISL1
|
ISL LIM homeobox 1 |
| chr3_-_133614597 | 0.06 |
ENST00000285208.4
ENST00000460865.3 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chrX_+_100353153 | 0.06 |
ENST00000423383.1
ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI
|
centromere protein I |
| chr3_+_32726620 | 0.06 |
ENST00000331889.6
ENST00000328834.5 |
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr21_-_27107283 | 0.06 |
ENST00000284971.3
ENST00000400099.1 |
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr16_+_77233294 | 0.06 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr19_+_17638059 | 0.06 |
ENST00000599164.1
ENST00000449408.2 ENST00000600871.1 ENST00000599124.1 |
FAM129C
|
family with sequence similarity 129, member C |
| chr1_+_15272271 | 0.06 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr6_-_97731019 | 0.06 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr3_-_125094093 | 0.06 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr2_-_216003127 | 0.06 |
ENST00000412081.1
ENST00000272895.7 |
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr4_-_186456652 | 0.06 |
ENST00000284767.5
ENST00000284770.5 |
PDLIM3
|
PDZ and LIM domain 3 |
| chr6_-_106773610 | 0.06 |
ENST00000369076.3
ENST00000369070.1 |
ATG5
|
autophagy related 5 |
| chr15_-_34502278 | 0.06 |
ENST00000559515.1
ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr2_+_149894968 | 0.06 |
ENST00000409642.3
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr12_+_54378849 | 0.05 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr5_+_85913721 | 0.05 |
ENST00000247655.3
ENST00000509578.1 ENST00000515763.1 |
COX7C
|
cytochrome c oxidase subunit VIIc |
| chr15_-_83474806 | 0.05 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr2_-_39664405 | 0.05 |
ENST00000341681.5
ENST00000263881.3 |
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr2_+_27498289 | 0.05 |
ENST00000296097.3
ENST00000420191.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr20_+_44637526 | 0.05 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr3_+_72201910 | 0.05 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr15_-_83621435 | 0.05 |
ENST00000450735.2
ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2
|
homer homolog 2 (Drosophila) |
| chr21_+_30502806 | 0.05 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr16_+_66429358 | 0.05 |
ENST00000539168.1
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr20_-_43438912 | 0.05 |
ENST00000541604.2
ENST00000372851.3 |
RIMS4
|
regulating synaptic membrane exocytosis 4 |
| chr1_+_113009163 | 0.05 |
ENST00000256640.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr1_-_204135450 | 0.05 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr21_-_27107344 | 0.05 |
ENST00000457143.2
|
ATP5J
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F6 |
| chr2_+_149895207 | 0.04 |
ENST00000409876.1
|
LYPD6B
|
LY6/PLAUR domain containing 6B |
| chr11_+_120081475 | 0.04 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr22_-_32341336 | 0.04 |
ENST00000248984.3
|
C22orf24
|
chromosome 22 open reading frame 24 |
| chr14_-_69619823 | 0.04 |
ENST00000341516.5
|
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr11_+_111749650 | 0.04 |
ENST00000528125.1
|
C11orf1
|
chromosome 11 open reading frame 1 |
| chr1_+_89150245 | 0.04 |
ENST00000370513.5
|
PKN2
|
protein kinase N2 |
| chr19_-_44174330 | 0.04 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr4_-_186456766 | 0.04 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr9_-_99637820 | 0.04 |
ENST00000289032.8
ENST00000535338.1 |
ZNF782
|
zinc finger protein 782 |
| chr3_-_52002403 | 0.04 |
ENST00000490063.1
ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4
|
poly(rC) binding protein 4 |
| chr15_-_34502197 | 0.04 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr14_-_69619689 | 0.04 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr12_-_90024360 | 0.04 |
ENST00000393164.2
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_+_54378923 | 0.04 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr2_+_122494676 | 0.03 |
ENST00000455432.1
|
TSN
|
translin |
| chr20_-_60942326 | 0.03 |
ENST00000370677.3
ENST00000370692.3 |
LAMA5
|
laminin, alpha 5 |
| chr2_-_197675000 | 0.03 |
ENST00000342506.2
|
C2orf66
|
chromosome 2 open reading frame 66 |
| chr2_+_233320827 | 0.03 |
ENST00000295463.3
|
ALPI
|
alkaline phosphatase, intestinal |
| chr14_-_74025625 | 0.03 |
ENST00000553558.1
ENST00000563329.1 ENST00000334988.2 ENST00000560393.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr17_-_56084578 | 0.03 |
ENST00000582730.2
ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1
|
serine/arginine-rich splicing factor 1 |
| chr5_-_176433350 | 0.03 |
ENST00000377227.4
ENST00000377219.2 |
UIMC1
|
ubiquitin interaction motif containing 1 |
| chrX_-_48216101 | 0.03 |
ENST00000298396.2
ENST00000376893.3 |
SSX3
|
synovial sarcoma, X breakpoint 3 |
| chrX_+_100663243 | 0.03 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr3_-_100712352 | 0.03 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr10_-_5060147 | 0.03 |
ENST00000604507.1
|
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr1_+_17516275 | 0.03 |
ENST00000412427.1
|
RP11-380J14.1
|
RP11-380J14.1 |
| chr14_-_69619291 | 0.03 |
ENST00000554215.1
ENST00000556847.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr3_-_100712292 | 0.03 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr10_+_4868460 | 0.03 |
ENST00000532248.1
ENST00000345253.5 ENST00000334019.4 |
AKR1E2
|
aldo-keto reductase family 1, member E2 |
| chr14_+_96722539 | 0.03 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr11_-_61197406 | 0.03 |
ENST00000541963.1
ENST00000477890.2 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr19_+_17638041 | 0.03 |
ENST00000601861.1
|
FAM129C
|
family with sequence similarity 129, member C |
| chr17_-_26662464 | 0.03 |
ENST00000579419.1
ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr5_-_131330272 | 0.03 |
ENST00000379240.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr3_+_133502877 | 0.03 |
ENST00000466490.2
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr10_-_76868866 | 0.03 |
ENST00000607487.1
|
DUSP13
|
dual specificity phosphatase 13 |
| chr3_+_14058794 | 0.02 |
ENST00000424053.1
ENST00000528067.1 ENST00000429201.1 |
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr2_+_162087577 | 0.02 |
ENST00000439442.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr11_-_64889252 | 0.02 |
ENST00000525297.1
ENST00000529259.1 |
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr20_+_34129770 | 0.02 |
ENST00000348547.2
ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3
|
ERGIC and golgi 3 |
| chr19_-_17958832 | 0.02 |
ENST00000458235.1
|
JAK3
|
Janus kinase 3 |
| chr16_-_88717482 | 0.02 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr2_-_241737128 | 0.02 |
ENST00000404283.3
|
KIF1A
|
kinesin family member 1A |
| chr14_-_94984181 | 0.02 |
ENST00000341228.2
|
SERPINA12
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| chr2_+_192543694 | 0.02 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr7_-_81399355 | 0.02 |
ENST00000457544.2
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_+_117864815 | 0.02 |
ENST00000433239.1
|
ANKRD7
|
ankyrin repeat domain 7 |
| chr10_-_5060201 | 0.02 |
ENST00000407674.1
|
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr17_+_57970469 | 0.02 |
ENST00000443572.2
ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| chr21_+_25801041 | 0.02 |
ENST00000453784.2
ENST00000423581.1 |
AP000476.1
|
AP000476.1 |
| chr1_+_27648709 | 0.02 |
ENST00000608611.1
ENST00000466759.1 ENST00000464813.1 ENST00000498220.1 |
TMEM222
|
transmembrane protein 222 |
| chr14_+_75746664 | 0.02 |
ENST00000557139.1
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr1_+_78245303 | 0.02 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chr11_-_44972476 | 0.02 |
ENST00000527685.1
ENST00000308212.5 |
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr5_-_131330315 | 0.02 |
ENST00000419502.1
|
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr1_-_224517823 | 0.02 |
ENST00000469968.1
ENST00000436927.1 ENST00000469075.1 ENST00000488718.1 ENST00000482491.1 ENST00000340871.4 ENST00000492281.1 ENST00000361463.3 ENST00000391875.2 ENST00000461546.1 |
NVL
|
nuclear VCP-like |
| chr11_-_8739383 | 0.02 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr6_-_130031358 | 0.02 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr4_+_158493642 | 0.02 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr5_-_137610254 | 0.02 |
ENST00000378362.3
|
GFRA3
|
GDNF family receptor alpha 3 |
| chr3_+_102153859 | 0.02 |
ENST00000306176.1
ENST00000466937.1 |
ZPLD1
|
zona pellucida-like domain containing 1 |
| chr8_-_30890710 | 0.02 |
ENST00000523392.1
|
PURG
|
purine-rich element binding protein G |
| chr6_+_83073334 | 0.02 |
ENST00000369750.3
|
TPBG
|
trophoblast glycoprotein |
| chr7_-_28220354 | 0.02 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr14_-_73997901 | 0.02 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr2_-_152146385 | 0.02 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr2_+_108905095 | 0.02 |
ENST00000251481.6
ENST00000326853.5 |
SULT1C2
|
sulfotransferase family, cytosolic, 1C, member 2 |
| chr7_+_110731062 | 0.02 |
ENST00000308478.5
ENST00000451085.1 ENST00000422987.3 ENST00000421101.1 |
LRRN3
|
leucine rich repeat neuronal 3 |
| chr20_-_31124186 | 0.01 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr12_-_122711968 | 0.01 |
ENST00000485724.1
|
DIABLO
|
diablo, IAP-binding mitochondrial protein |
| chr16_+_67313412 | 0.01 |
ENST00000379344.3
ENST00000568621.1 ENST00000450733.1 ENST00000567938.1 |
PLEKHG4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr6_-_33679452 | 0.01 |
ENST00000374231.4
ENST00000607484.1 ENST00000374214.3 |
UQCC2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr12_-_52867569 | 0.01 |
ENST00000252250.6
|
KRT6C
|
keratin 6C |
| chr10_-_23003460 | 0.01 |
ENST00000376573.4
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
| chr1_+_89149905 | 0.01 |
ENST00000316005.7
ENST00000370521.3 ENST00000370505.3 |
PKN2
|
protein kinase N2 |
| chr12_-_49351303 | 0.01 |
ENST00000256682.4
|
ARF3
|
ADP-ribosylation factor 3 |
| chr22_-_41215291 | 0.01 |
ENST00000542412.1
ENST00000544408.1 |
SLC25A17
|
solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 |
| chr10_+_51565188 | 0.01 |
ENST00000430396.2
ENST00000374087.4 ENST00000414907.2 |
NCOA4
|
nuclear receptor coactivator 4 |
| chr2_+_166152283 | 0.01 |
ENST00000375427.2
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr4_+_114066764 | 0.01 |
ENST00000511380.1
|
ANK2
|
ankyrin 2, neuronal |
| chr5_+_139554227 | 0.01 |
ENST00000261811.4
|
CYSTM1
|
cysteine-rich transmembrane module containing 1 |
| chr1_+_203765437 | 0.01 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr19_+_7895074 | 0.01 |
ENST00000270530.4
|
EVI5L
|
ecotropic viral integration site 5-like |
| chr7_+_134576317 | 0.01 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr7_-_81399438 | 0.01 |
ENST00000222390.5
|
HGF
|
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr10_-_115423792 | 0.01 |
ENST00000369360.3
ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP
|
nebulin-related anchoring protein |
| chr1_+_39456895 | 0.01 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr5_+_67576109 | 0.01 |
ENST00000522084.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr16_+_69221028 | 0.00 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr2_+_27498331 | 0.00 |
ENST00000402462.1
ENST00000404433.1 ENST00000406962.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr19_+_58258164 | 0.00 |
ENST00000317178.5
|
ZNF776
|
zinc finger protein 776 |
| chr21_+_27107672 | 0.00 |
ENST00000400075.3
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa |
| chr12_-_56710118 | 0.00 |
ENST00000273308.4
|
CNPY2
|
canopy FGF signaling regulator 2 |
| chr1_-_26633067 | 0.00 |
ENST00000421827.2
ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11
|
UBX domain protein 11 |
| chr7_-_229557 | 0.00 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr20_+_32951070 | 0.00 |
ENST00000535650.1
ENST00000262650.6 |
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr5_-_131329918 | 0.00 |
ENST00000357096.1
ENST00000431707.1 ENST00000434099.1 |
ACSL6
|
acyl-CoA synthetase long-chain family member 6 |
| chr20_+_32951041 | 0.00 |
ENST00000374864.4
|
ITCH
|
itchy E3 ubiquitin protein ligase |
| chr22_+_19929130 | 0.00 |
ENST00000361682.6
ENST00000403184.1 ENST00000403710.1 ENST00000407537.1 |
COMT
|
catechol-O-methyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of T
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 0.5 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.1 | 0.4 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.2 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.4 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.2 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.2 | GO:0044034 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0048936 | visceral motor neuron differentiation(GO:0021524) peripheral nervous system neuron axonogenesis(GO:0048936) cardiac cell fate determination(GO:0060913) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.2 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |