Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
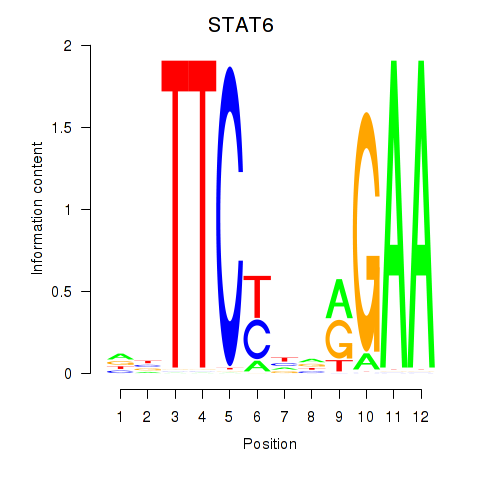
Results for STAT6
Z-value: 0.54
Transcription factors associated with STAT6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT6
|
ENSG00000166888.6 | signal transducer and activator of transcription 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT6 | hg19_v2_chr12_-_57504069_57504123 | 0.45 | 3.7e-01 | Click! |
Activity profile of STAT6 motif
Sorted Z-values of STAT6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_172034218 | 0.61 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr17_+_61473104 | 0.28 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr11_-_14379997 | 0.27 |
ENST00000526063.1
ENST00000532814.1 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr1_+_45965725 | 0.24 |
ENST00000401061.4
|
MMACHC
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr6_+_138188378 | 0.22 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr14_-_24733444 | 0.21 |
ENST00000560478.1
ENST00000560443.1 |
TGM1
|
transglutaminase 1 |
| chr5_+_35856951 | 0.21 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr9_-_3469181 | 0.20 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr7_+_106415457 | 0.20 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr7_-_38969150 | 0.19 |
ENST00000418457.2
|
VPS41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr6_-_41254403 | 0.19 |
ENST00000589614.1
ENST00000334475.6 ENST00000591620.1 ENST00000244709.4 |
TREM1
|
triggering receptor expressed on myeloid cells 1 |
| chr18_+_66382428 | 0.19 |
ENST00000578970.1
ENST00000582371.1 ENST00000584775.1 |
CCDC102B
|
coiled-coil domain containing 102B |
| chrX_+_135251783 | 0.18 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr11_+_73358594 | 0.17 |
ENST00000227214.6
ENST00000398494.4 ENST00000543085.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr5_+_102201687 | 0.17 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chrX_+_135252050 | 0.16 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr2_+_191792376 | 0.15 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr2_-_120124258 | 0.15 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr9_-_100000957 | 0.15 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr14_+_55034599 | 0.15 |
ENST00000392067.3
ENST00000357634.3 |
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr12_+_6493319 | 0.15 |
ENST00000536876.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr18_-_21017817 | 0.14 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr6_+_138188351 | 0.14 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr13_+_73629107 | 0.13 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr5_+_102201722 | 0.13 |
ENST00000274392.9
ENST00000455264.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chrX_+_135251835 | 0.13 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr6_+_30951487 | 0.12 |
ENST00000486149.2
ENST00000376296.3 |
MUC21
|
mucin 21, cell surface associated |
| chr12_+_6493406 | 0.12 |
ENST00000543190.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr5_+_93954358 | 0.11 |
ENST00000504099.1
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr2_-_120124383 | 0.11 |
ENST00000334816.7
|
C2orf76
|
chromosome 2 open reading frame 76 |
| chr17_+_40912764 | 0.11 |
ENST00000589683.1
ENST00000588928.1 |
RAMP2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr10_+_124907638 | 0.11 |
ENST00000339992.3
|
HMX2
|
H6 family homeobox 2 |
| chr7_+_112120908 | 0.11 |
ENST00000439068.2
ENST00000312849.4 ENST00000429049.1 |
LSMEM1
|
leucine-rich single-pass membrane protein 1 |
| chr11_-_119252359 | 0.11 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr2_+_197504278 | 0.11 |
ENST00000272831.7
ENST00000389175.4 ENST00000472405.2 ENST00000423093.2 |
CCDC150
|
coiled-coil domain containing 150 |
| chr1_+_66796401 | 0.11 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr16_+_67694849 | 0.10 |
ENST00000602551.1
ENST00000458121.2 ENST00000219255.3 |
PARD6A
|
par-6 family cell polarity regulator alpha |
| chr7_+_23637763 | 0.10 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr3_+_52448539 | 0.10 |
ENST00000461861.1
|
PHF7
|
PHD finger protein 7 |
| chr11_+_65082289 | 0.10 |
ENST00000279249.2
|
CDC42EP2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chrX_+_102192200 | 0.10 |
ENST00000218249.5
|
RAB40AL
|
RAB40A, member RAS oncogene family-like |
| chr14_+_56127960 | 0.10 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr12_+_6493199 | 0.10 |
ENST00000228918.4
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr14_-_24551195 | 0.10 |
ENST00000560550.1
|
NRL
|
neural retina leucine zipper |
| chr1_+_45274154 | 0.10 |
ENST00000450269.1
ENST00000453418.1 ENST00000409335.2 |
BTBD19
|
BTB (POZ) domain containing 19 |
| chr5_+_112227311 | 0.10 |
ENST00000391338.1
|
ZRSR1
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr17_-_34207295 | 0.10 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr14_-_24551137 | 0.09 |
ENST00000396995.1
|
NRL
|
neural retina leucine zipper |
| chr1_-_8086343 | 0.09 |
ENST00000474874.1
ENST00000469499.1 ENST00000377482.5 |
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr6_+_31887761 | 0.09 |
ENST00000413154.1
|
C2
|
complement component 2 |
| chr1_+_32608566 | 0.09 |
ENST00000545542.1
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7) |
| chr16_+_69984810 | 0.09 |
ENST00000393701.2
ENST00000568461.1 |
CLEC18A
|
C-type lectin domain family 18, member A |
| chr15_+_85525205 | 0.09 |
ENST00000394553.1
ENST00000339708.5 |
PDE8A
|
phosphodiesterase 8A |
| chr6_-_30524951 | 0.09 |
ENST00000376621.3
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr2_-_175462456 | 0.08 |
ENST00000409891.1
ENST00000410117.1 |
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr14_+_56127989 | 0.08 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr14_-_46185155 | 0.08 |
ENST00000555442.1
|
RP11-369C8.1
|
RP11-369C8.1 |
| chr8_+_107593198 | 0.08 |
ENST00000517686.1
|
OXR1
|
oxidation resistance 1 |
| chr15_+_96869165 | 0.08 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr12_+_26164645 | 0.08 |
ENST00000542004.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr3_-_37218023 | 0.08 |
ENST00000416425.1
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr22_-_31364187 | 0.07 |
ENST00000215862.4
ENST00000397641.3 |
MORC2
|
MORC family CW-type zinc finger 2 |
| chr3_+_189507460 | 0.07 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr7_+_76091775 | 0.07 |
ENST00000442516.1
|
DTX2
|
deltex homolog 2 (Drosophila) |
| chr11_-_77531752 | 0.07 |
ENST00000440064.2
ENST00000528095.1 |
RSF1
|
remodeling and spacing factor 1 |
| chr12_+_9144626 | 0.07 |
ENST00000543895.1
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1 |
| chr3_+_10312604 | 0.07 |
ENST00000426850.1
|
TATDN2
|
TatD DNase domain containing 2 |
| chr1_+_82165350 | 0.07 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chrX_-_138914394 | 0.07 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr2_-_190044480 | 0.07 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr20_+_42574317 | 0.07 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr18_-_24445664 | 0.07 |
ENST00000578776.1
|
AQP4
|
aquaporin 4 |
| chr11_-_119252425 | 0.06 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr15_+_27111510 | 0.06 |
ENST00000335625.5
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr7_+_138818490 | 0.06 |
ENST00000430935.1
ENST00000495038.1 ENST00000474035.2 ENST00000478836.2 ENST00000464848.1 ENST00000343187.4 |
TTC26
|
tetratricopeptide repeat domain 26 |
| chr16_-_29757272 | 0.06 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr11_+_65101225 | 0.06 |
ENST00000528416.1
ENST00000415073.2 ENST00000252268.4 |
DPF2
|
D4, zinc and double PHD fingers family 2 |
| chr1_-_41328018 | 0.06 |
ENST00000372638.2
|
CITED4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr3_+_189507523 | 0.06 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr4_+_141677577 | 0.06 |
ENST00000609937.1
|
RP11-102N12.3
|
RP11-102N12.3 |
| chr12_-_53097247 | 0.06 |
ENST00000341809.3
ENST00000537195.1 |
KRT77
|
keratin 77 |
| chr9_+_90112767 | 0.06 |
ENST00000408954.3
|
DAPK1
|
death-associated protein kinase 1 |
| chr9_+_36572851 | 0.06 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr21_+_17792672 | 0.06 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr2_+_103378472 | 0.06 |
ENST00000412401.2
|
TMEM182
|
transmembrane protein 182 |
| chr9_+_90112741 | 0.05 |
ENST00000469640.2
|
DAPK1
|
death-associated protein kinase 1 |
| chr17_+_32582293 | 0.05 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr19_-_54604083 | 0.05 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr1_-_36020531 | 0.05 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr9_+_90112590 | 0.05 |
ENST00000472284.1
|
DAPK1
|
death-associated protein kinase 1 |
| chr3_+_121311966 | 0.05 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr8_-_117886732 | 0.05 |
ENST00000517485.1
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr12_-_54779511 | 0.05 |
ENST00000551109.1
ENST00000546970.1 |
ZNF385A
|
zinc finger protein 385A |
| chr8_+_104311059 | 0.05 |
ENST00000358755.4
ENST00000523739.1 ENST00000540287.1 |
FZD6
|
frizzled family receptor 6 |
| chr16_-_74455290 | 0.05 |
ENST00000339953.5
|
CLEC18B
|
C-type lectin domain family 18, member B |
| chr2_+_234526272 | 0.05 |
ENST00000373450.4
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr9_+_90112117 | 0.05 |
ENST00000358077.5
|
DAPK1
|
death-associated protein kinase 1 |
| chr6_-_11232891 | 0.05 |
ENST00000379433.5
ENST00000379446.5 |
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr11_+_118230287 | 0.05 |
ENST00000252108.3
ENST00000431736.2 |
UBE4A
|
ubiquitination factor E4A |
| chr15_+_101402041 | 0.05 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr7_+_129015671 | 0.05 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr16_-_67694597 | 0.05 |
ENST00000393919.4
ENST00000219251.8 |
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chrX_-_73511908 | 0.04 |
ENST00000455395.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr2_+_113885138 | 0.04 |
ENST00000409930.3
|
IL1RN
|
interleukin 1 receptor antagonist |
| chr6_+_30525051 | 0.04 |
ENST00000376557.3
|
PRR3
|
proline rich 3 |
| chr2_-_204400013 | 0.04 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr6_-_32821599 | 0.04 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr1_+_61869748 | 0.04 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr9_-_135996537 | 0.04 |
ENST00000372050.3
ENST00000372047.3 |
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr17_-_39681578 | 0.04 |
ENST00000593096.1
|
KRT19
|
keratin 19 |
| chr15_-_64385981 | 0.04 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr2_-_31440377 | 0.04 |
ENST00000444918.2
ENST00000403897.3 |
CAPN14
|
calpain 14 |
| chr22_+_38201114 | 0.04 |
ENST00000340857.2
|
H1F0
|
H1 histone family, member 0 |
| chr4_-_46391931 | 0.04 |
ENST00000381620.4
|
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr13_+_53226963 | 0.04 |
ENST00000343788.6
ENST00000535397.1 ENST00000310528.8 |
SUGT1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr12_-_42877726 | 0.04 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr13_-_28545276 | 0.04 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr4_+_144312659 | 0.04 |
ENST00000509992.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr12_-_6451235 | 0.04 |
ENST00000440083.2
ENST00000162749.2 |
TNFRSF1A
|
tumor necrosis factor receptor superfamily, member 1A |
| chr2_+_219745020 | 0.04 |
ENST00000258411.3
|
WNT10A
|
wingless-type MMTV integration site family, member 10A |
| chr16_+_82090028 | 0.04 |
ENST00000568090.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr11_+_77532155 | 0.03 |
ENST00000532481.1
ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr3_+_132379154 | 0.03 |
ENST00000468022.1
ENST00000473651.1 ENST00000494238.2 |
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chr11_-_128457446 | 0.03 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr17_-_62308087 | 0.03 |
ENST00000583097.1
|
TEX2
|
testis expressed 2 |
| chr21_+_42539701 | 0.03 |
ENST00000330333.6
ENST00000328735.6 ENST00000347667.5 |
BACE2
|
beta-site APP-cleaving enzyme 2 |
| chr1_-_205649580 | 0.03 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr8_-_37594944 | 0.03 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr14_-_37641618 | 0.03 |
ENST00000555449.1
|
SLC25A21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr2_-_230786619 | 0.03 |
ENST00000389045.3
ENST00000409677.1 |
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_-_49851313 | 0.03 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr6_+_26440700 | 0.03 |
ENST00000494393.1
ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3
|
butyrophilin, subfamily 3, member A3 |
| chr9_+_131464767 | 0.03 |
ENST00000291906.4
|
PKN3
|
protein kinase N3 |
| chr10_-_61122220 | 0.03 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr20_-_5931051 | 0.03 |
ENST00000453074.2
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr15_+_25200074 | 0.03 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chr12_-_42877764 | 0.03 |
ENST00000455697.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr2_+_131769256 | 0.03 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr1_-_45965525 | 0.03 |
ENST00000488405.2
ENST00000490551.3 ENST00000432082.1 |
CCDC163P
|
coiled-coil domain containing 163, pseudogene |
| chr6_+_31730773 | 0.03 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr2_-_230786032 | 0.03 |
ENST00000428959.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr9_-_10612703 | 0.02 |
ENST00000463477.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr11_-_67981046 | 0.02 |
ENST00000402789.1
ENST00000402185.2 ENST00000458496.1 |
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr14_-_23285069 | 0.02 |
ENST00000554758.1
ENST00000397528.4 |
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr2_-_133427767 | 0.02 |
ENST00000397463.2
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr11_-_115158193 | 0.02 |
ENST00000543540.1
|
CADM1
|
cell adhesion molecule 1 |
| chr6_+_32821924 | 0.02 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr14_-_23285011 | 0.02 |
ENST00000397532.3
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr17_-_60005365 | 0.02 |
ENST00000444766.3
|
INTS2
|
integrator complex subunit 2 |
| chr12_-_51402984 | 0.02 |
ENST00000545993.2
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr4_+_166300084 | 0.02 |
ENST00000402744.4
|
CPE
|
carboxypeptidase E |
| chr11_-_77531858 | 0.02 |
ENST00000360355.2
|
RSF1
|
remodeling and spacing factor 1 |
| chr11_-_10828892 | 0.02 |
ENST00000525681.1
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr1_+_202431859 | 0.02 |
ENST00000391959.3
ENST00000367270.4 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr2_+_202316392 | 0.02 |
ENST00000194530.3
ENST00000392249.2 |
STRADB
|
STE20-related kinase adaptor beta |
| chr6_+_147527103 | 0.02 |
ENST00000179882.6
|
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr13_-_45915221 | 0.02 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr16_+_30075463 | 0.02 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr19_+_13228917 | 0.02 |
ENST00000586171.1
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr1_-_241803679 | 0.02 |
ENST00000331838.5
|
OPN3
|
opsin 3 |
| chr3_+_189507432 | 0.02 |
ENST00000354600.5
|
TP63
|
tumor protein p63 |
| chr16_-_67753206 | 0.02 |
ENST00000268797.7
|
GFOD2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr17_+_7461580 | 0.02 |
ENST00000483039.1
ENST00000396542.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr10_-_35379241 | 0.01 |
ENST00000374748.1
ENST00000374749.3 |
CUL2
|
cullin 2 |
| chr17_+_68071389 | 0.01 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr3_-_120365866 | 0.01 |
ENST00000475447.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr2_+_182756615 | 0.01 |
ENST00000431877.2
ENST00000320370.7 |
SSFA2
|
sperm specific antigen 2 |
| chr17_+_28268623 | 0.01 |
ENST00000394835.3
ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr19_+_13229126 | 0.01 |
ENST00000292431.4
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr5_+_118691706 | 0.01 |
ENST00000415806.2
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr1_+_26759295 | 0.01 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr15_+_30918879 | 0.01 |
ENST00000428041.2
|
ARHGAP11B
|
Rho GTPase activating protein 11B |
| chr15_-_64386120 | 0.01 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr3_-_129513259 | 0.01 |
ENST00000329333.5
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr22_+_44464923 | 0.01 |
ENST00000404989.1
|
PARVB
|
parvin, beta |
| chr15_+_64386261 | 0.01 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chr15_+_25200108 | 0.01 |
ENST00000577949.1
ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF
SNRPN
|
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr17_-_34890037 | 0.01 |
ENST00000589404.1
|
MYO19
|
myosin XIX |
| chr3_-_98235962 | 0.01 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr2_-_26700900 | 0.01 |
ENST00000338581.6
ENST00000339598.3 ENST00000402415.3 |
OTOF
|
otoferlin |
| chr6_+_84222220 | 0.01 |
ENST00000369700.3
|
PRSS35
|
protease, serine, 35 |
| chr2_+_182756915 | 0.01 |
ENST00000428267.2
|
SSFA2
|
sperm specific antigen 2 |
| chr11_+_7618413 | 0.01 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr4_+_30721968 | 0.01 |
ENST00000361762.2
|
PCDH7
|
protocadherin 7 |
| chr1_-_108735440 | 0.01 |
ENST00000370041.4
|
SLC25A24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr17_-_34890709 | 0.01 |
ENST00000544606.1
|
MYO19
|
myosin XIX |
| chr1_+_89246647 | 0.01 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr16_+_30076052 | 0.01 |
ENST00000563987.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr18_-_53068782 | 0.01 |
ENST00000569012.1
|
TCF4
|
transcription factor 4 |
| chr2_-_204399976 | 0.01 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chrX_-_100546314 | 0.01 |
ENST00000356784.1
|
TAF7L
|
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr15_-_88799661 | 0.01 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr3_+_153839149 | 0.01 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr2_-_16804320 | 0.01 |
ENST00000355549.2
|
FAM49A
|
family with sequence similarity 49, member A |
| chr6_-_27860956 | 0.01 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr1_-_201915590 | 0.01 |
ENST00000367288.4
|
LMOD1
|
leiomodin 1 (smooth muscle) |
| chr2_+_234686976 | 0.01 |
ENST00000389758.3
|
MROH2A
|
maestro heat-like repeat family member 2A |
| chr14_-_24553834 | 0.00 |
ENST00000397002.2
|
NRL
|
neural retina leucine zipper |
| chr14_+_61449076 | 0.00 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr5_+_138609441 | 0.00 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070429 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.1 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.2 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.2 | GO:0070945 | neutrophil mediated killing of gram-negative bacterium(GO:0070945) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.2 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.0 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 0.0 | GO:0046967 | cytosol to ER transport(GO:0046967) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.1 | 0.3 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.0 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |