Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
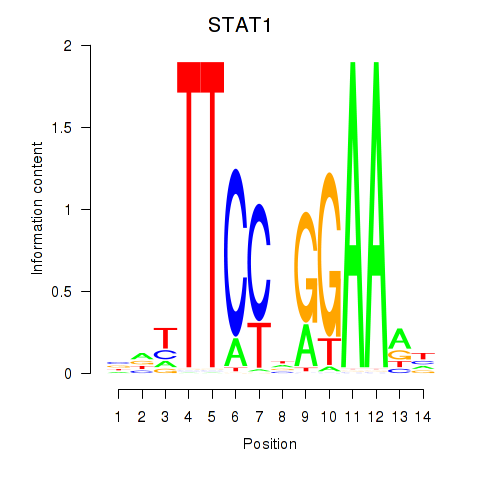
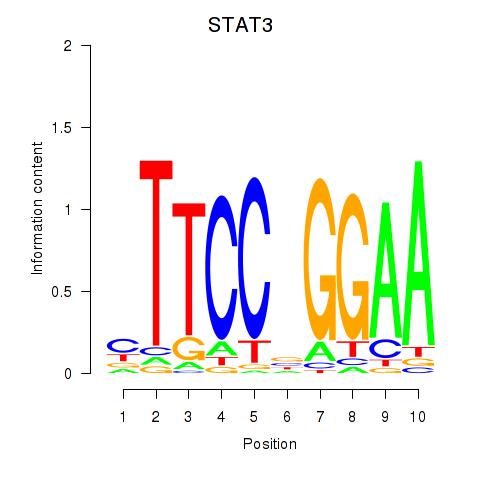
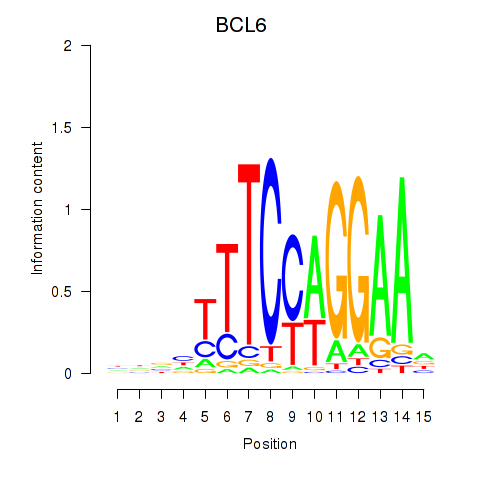
Results for STAT1_STAT3_BCL6
Z-value: 0.97
Transcription factors associated with STAT1_STAT3_BCL6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT1
|
ENSG00000115415.14 | signal transducer and activator of transcription 1 |
|
STAT3
|
ENSG00000168610.10 | signal transducer and activator of transcription 3 |
|
BCL6
|
ENSG00000113916.13 | BCL6 transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BCL6 | hg19_v2_chr3_-_187455680_187455732 | -0.53 | 2.8e-01 | Click! |
| STAT3 | hg19_v2_chr17_-_40540586_40540600 | -0.33 | 5.2e-01 | Click! |
| STAT1 | hg19_v2_chr2_-_191878874_191878976 | 0.15 | 7.8e-01 | Click! |
Activity profile of STAT1_STAT3_BCL6 motif
Sorted Z-values of STAT1_STAT3_BCL6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_50649192 | 1.94 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr17_-_46692457 | 1.81 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr19_-_4717835 | 0.97 |
ENST00000599248.1
|
DPP9
|
dipeptidyl-peptidase 9 |
| chr3_-_52864680 | 0.76 |
ENST00000406595.1
ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr16_-_67517716 | 0.68 |
ENST00000290953.2
|
AGRP
|
agouti related protein homolog (mouse) |
| chr6_+_26104104 | 0.67 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr2_+_9778872 | 0.65 |
ENST00000478468.1
|
RP11-521D12.1
|
RP11-521D12.1 |
| chr17_+_2264983 | 0.62 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr12_-_42878101 | 0.61 |
ENST00000552108.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr3_-_145940214 | 0.56 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr17_-_79917645 | 0.56 |
ENST00000477214.1
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
| chr9_-_134151915 | 0.56 |
ENST00000372271.3
|
FAM78A
|
family with sequence similarity 78, member A |
| chr9_-_117880477 | 0.51 |
ENST00000534839.1
ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC
|
tenascin C |
| chr5_-_38845812 | 0.45 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr11_+_111412271 | 0.41 |
ENST00000528102.1
|
LAYN
|
layilin |
| chr3_+_112930946 | 0.40 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr2_+_17935119 | 0.38 |
ENST00000317402.7
|
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr12_-_48213735 | 0.37 |
ENST00000417902.1
ENST00000417107.1 |
HDAC7
|
histone deacetylase 7 |
| chr19_+_57106624 | 0.37 |
ENST00000599599.1
|
ZNF71
|
zinc finger protein 71 |
| chr17_+_76356516 | 0.36 |
ENST00000592569.1
|
RP11-806H10.4
|
RP11-806H10.4 |
| chr19_+_35225060 | 0.35 |
ENST00000599244.1
ENST00000392232.3 |
ZNF181
|
zinc finger protein 181 |
| chr19_+_15783879 | 0.34 |
ENST00000551607.1
|
CYP4F12
|
cytochrome P450, family 4, subfamily F, polypeptide 12 |
| chr10_+_35484793 | 0.34 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr13_-_34250861 | 0.34 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr22_-_39637135 | 0.33 |
ENST00000440375.1
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr16_+_14396121 | 0.32 |
ENST00000570945.1
|
RP11-65J21.3
|
RP11-65J21.3 |
| chr12_+_56414851 | 0.32 |
ENST00000547167.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr19_+_35168633 | 0.32 |
ENST00000505365.2
|
ZNF302
|
zinc finger protein 302 |
| chr12_-_7245018 | 0.31 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr6_+_131571535 | 0.31 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr1_+_155006300 | 0.30 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr10_+_72194585 | 0.29 |
ENST00000420338.2
|
AC022532.1
|
Uncharacterized protein |
| chr6_+_84222220 | 0.29 |
ENST00000369700.3
|
PRSS35
|
protease, serine, 35 |
| chr2_+_87754989 | 0.29 |
ENST00000409898.2
ENST00000419680.2 ENST00000414584.1 ENST00000455131.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr6_-_18264706 | 0.28 |
ENST00000244776.7
ENST00000503715.1 |
DEK
|
DEK oncogene |
| chr2_+_87755054 | 0.28 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr19_-_56904799 | 0.28 |
ENST00000589895.1
ENST00000589143.1 ENST00000301310.4 ENST00000586929.1 |
ZNF582
|
zinc finger protein 582 |
| chr12_-_133050726 | 0.28 |
ENST00000595994.1
|
MUC8
|
mucin 8 |
| chr16_+_30934376 | 0.27 |
ENST00000562798.1
ENST00000471231.2 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr1_+_16083098 | 0.27 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr16_+_48657361 | 0.27 |
ENST00000565072.1
|
RP11-42I10.1
|
RP11-42I10.1 |
| chr19_+_10197463 | 0.26 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr3_+_122399697 | 0.26 |
ENST00000494811.1
|
PARP14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr12_+_56325231 | 0.25 |
ENST00000549368.1
|
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr6_+_292253 | 0.25 |
ENST00000603453.1
ENST00000605315.1 ENST00000603881.1 |
DUSP22
|
dual specificity phosphatase 22 |
| chr12_-_7244469 | 0.25 |
ENST00000538050.1
ENST00000536053.2 |
C1R
|
complement component 1, r subcomponent |
| chr12_+_56324933 | 0.24 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr8_-_102803163 | 0.24 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr4_-_155511887 | 0.24 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr3_-_148939598 | 0.24 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr22_-_31536480 | 0.23 |
ENST00000215885.3
|
PLA2G3
|
phospholipase A2, group III |
| chrX_+_106045891 | 0.23 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr5_-_111092930 | 0.23 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr1_+_59250815 | 0.23 |
ENST00000544621.1
ENST00000419531.2 |
RP4-794H19.2
|
long intergenic non-protein coding RNA 1135 |
| chr6_+_159290917 | 0.23 |
ENST00000367072.1
|
C6orf99
|
chromosome 6 open reading frame 99 |
| chr19_+_35168567 | 0.23 |
ENST00000457781.2
ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302
|
zinc finger protein 302 |
| chr12_+_56414795 | 0.23 |
ENST00000431367.2
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr12_-_11175219 | 0.22 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr4_+_86396321 | 0.21 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr16_-_74734672 | 0.21 |
ENST00000306247.7
ENST00000575686.1 |
MLKL
|
mixed lineage kinase domain-like |
| chr9_+_70971815 | 0.21 |
ENST00000396392.1
ENST00000396396.1 |
PGM5
|
phosphoglucomutase 5 |
| chr12_-_10007448 | 0.21 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr1_+_26759295 | 0.21 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr6_-_45544507 | 0.21 |
ENST00000563807.1
|
RP1-166H4.2
|
RP1-166H4.2 |
| chr3_-_4793274 | 0.20 |
ENST00000414938.1
|
EGOT
|
eosinophil granule ontogeny transcript (non-protein coding) |
| chr11_+_71710648 | 0.20 |
ENST00000260049.5
|
IL18BP
|
interleukin 18 binding protein |
| chr19_-_10613862 | 0.20 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr19_-_10613361 | 0.19 |
ENST00000591039.1
ENST00000591419.1 |
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr16_+_72088376 | 0.19 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr11_+_71710973 | 0.19 |
ENST00000393707.4
|
IL18BP
|
interleukin 18 binding protein |
| chr12_-_42877726 | 0.18 |
ENST00000548696.1
|
PRICKLE1
|
prickle homolog 1 (Drosophila) |
| chr2_-_204398141 | 0.18 |
ENST00000428637.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr16_-_90096309 | 0.18 |
ENST00000408886.2
|
C16orf3
|
chromosome 16 open reading frame 3 |
| chr21_-_34915123 | 0.18 |
ENST00000438059.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr10_+_6625733 | 0.18 |
ENST00000607982.1
ENST00000608526.1 |
PRKCQ-AS1
|
PRKCQ antisense RNA 1 |
| chr10_+_81065975 | 0.18 |
ENST00000446377.2
|
ZMIZ1
|
zinc finger, MIZ-type containing 1 |
| chr2_-_17935059 | 0.18 |
ENST00000448223.2
ENST00000381272.4 ENST00000351948.4 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr19_-_44405941 | 0.18 |
ENST00000587128.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr8_-_82645082 | 0.18 |
ENST00000523361.1
|
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr8_-_42358742 | 0.17 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr5_+_40841410 | 0.17 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr1_+_40997233 | 0.17 |
ENST00000372699.3
ENST00000372697.3 ENST00000372696.3 |
ZNF684
|
zinc finger protein 684 |
| chr15_+_71185148 | 0.17 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr10_-_99771079 | 0.17 |
ENST00000309155.3
|
CRTAC1
|
cartilage acidic protein 1 |
| chr19_-_4540486 | 0.17 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr19_+_10381769 | 0.17 |
ENST00000423829.2
ENST00000588645.1 |
ICAM1
|
intercellular adhesion molecule 1 |
| chr17_-_34207295 | 0.17 |
ENST00000463941.1
ENST00000293272.3 |
CCL5
|
chemokine (C-C motif) ligand 5 |
| chr9_-_117853297 | 0.17 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr20_-_18774614 | 0.17 |
ENST00000412553.1
|
LINC00652
|
long intergenic non-protein coding RNA 652 |
| chr2_-_165630264 | 0.17 |
ENST00000452626.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr12_+_58138664 | 0.17 |
ENST00000257910.3
|
TSPAN31
|
tetraspanin 31 |
| chr1_+_42619070 | 0.16 |
ENST00000372581.1
|
GUCA2B
|
guanylate cyclase activator 2B (uroguanylin) |
| chr19_+_56905024 | 0.16 |
ENST00000591172.1
ENST00000589888.1 ENST00000587979.1 ENST00000585659.1 ENST00000593109.1 |
ZNF582-AS1
|
ZNF582 antisense RNA 1 (head to head) |
| chr17_-_78428487 | 0.16 |
ENST00000562672.2
|
CTD-2526A2.2
|
CTD-2526A2.2 |
| chr17_+_42385927 | 0.16 |
ENST00000426726.3
ENST00000590941.1 ENST00000225441.7 |
RUNDC3A
|
RUN domain containing 3A |
| chr6_+_31730773 | 0.16 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr11_-_62609281 | 0.16 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr6_+_63921399 | 0.16 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr6_+_32821924 | 0.16 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr3_+_136537911 | 0.15 |
ENST00000393079.3
|
SLC35G2
|
solute carrier family 35, member G2 |
| chr7_-_54826869 | 0.15 |
ENST00000450622.1
|
SEC61G
|
Sec61 gamma subunit |
| chr17_+_74729060 | 0.15 |
ENST00000587459.1
|
RP11-318A15.7
|
Uncharacterized protein |
| chr21_+_34602377 | 0.15 |
ENST00000342101.3
ENST00000413881.1 ENST00000443073.1 |
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr22_-_37640456 | 0.15 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr5_-_140013241 | 0.15 |
ENST00000519715.1
|
CD14
|
CD14 molecule |
| chr6_+_44126545 | 0.15 |
ENST00000532171.1
ENST00000398776.1 ENST00000542245.1 |
CAPN11
|
calpain 11 |
| chr14_-_38036271 | 0.15 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr3_+_62304712 | 0.15 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr17_+_19030782 | 0.15 |
ENST00000344415.4
ENST00000577213.1 |
GRAPL
|
GRB2-related adaptor protein-like |
| chr14_-_50506589 | 0.14 |
ENST00000553914.2
|
RP11-58E21.3
|
RP11-58E21.3 |
| chr1_-_2706236 | 0.14 |
ENST00000401095.3
ENST00000574621.2 |
TTC34
|
tetratricopeptide repeat domain 34 |
| chr19_-_11688260 | 0.14 |
ENST00000590832.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr3_+_45430105 | 0.14 |
ENST00000430399.1
|
LARS2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr17_-_42994283 | 0.13 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr3_-_160117035 | 0.13 |
ENST00000489004.1
ENST00000496589.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr11_-_65363259 | 0.13 |
ENST00000342202.4
|
KCNK7
|
potassium channel, subfamily K, member 7 |
| chr14_+_24407940 | 0.13 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr22_-_39636914 | 0.13 |
ENST00000381551.4
|
PDGFB
|
platelet-derived growth factor beta polypeptide |
| chr3_+_62304648 | 0.13 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chrX_-_20237059 | 0.13 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr19_+_40503013 | 0.13 |
ENST00000595225.1
|
ZNF546
|
zinc finger protein 546 |
| chr17_-_26697304 | 0.13 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr10_-_6622201 | 0.13 |
ENST00000539722.1
ENST00000397176.2 |
PRKCQ
|
protein kinase C, theta |
| chr6_+_44194762 | 0.13 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr19_+_40502938 | 0.12 |
ENST00000599504.1
ENST00000596894.1 ENST00000601138.1 ENST00000600094.1 ENST00000347077.4 |
ZNF546
|
zinc finger protein 546 |
| chr2_-_96926313 | 0.12 |
ENST00000435268.1
|
TMEM127
|
transmembrane protein 127 |
| chr11_-_82997013 | 0.12 |
ENST00000529073.1
ENST00000529611.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr14_-_92414294 | 0.12 |
ENST00000554468.1
|
FBLN5
|
fibulin 5 |
| chr6_+_292051 | 0.12 |
ENST00000344450.5
|
DUSP22
|
dual specificity phosphatase 22 |
| chr1_-_153521597 | 0.12 |
ENST00000368712.1
|
S100A3
|
S100 calcium binding protein A3 |
| chr7_-_54826920 | 0.12 |
ENST00000395535.3
ENST00000352861.4 |
SEC61G
|
Sec61 gamma subunit |
| chr3_-_93692681 | 0.12 |
ENST00000348974.4
|
PROS1
|
protein S (alpha) |
| chr14_+_45605157 | 0.12 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr17_-_19265855 | 0.12 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr8_-_7320974 | 0.12 |
ENST00000528943.1
ENST00000359758.5 ENST00000361111.2 ENST00000398462.2 ENST00000297498.2 ENST00000317900.5 |
SPAG11B
|
sperm associated antigen 11B |
| chr12_-_11184006 | 0.12 |
ENST00000390675.2
|
TAS2R31
|
taste receptor, type 2, member 31 |
| chr16_-_20338748 | 0.12 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr5_+_169758393 | 0.12 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr14_+_45605127 | 0.12 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr5_-_145214893 | 0.12 |
ENST00000394450.2
|
PRELID2
|
PRELI domain containing 2 |
| chr10_+_129785536 | 0.11 |
ENST00000419012.2
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr4_+_86396265 | 0.11 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr4_+_41937131 | 0.11 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr9_+_136287444 | 0.11 |
ENST00000355699.2
ENST00000356589.2 ENST00000371911.3 |
ADAMTS13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr3_+_45067659 | 0.11 |
ENST00000296130.4
|
CLEC3B
|
C-type lectin domain family 3, member B |
| chr1_+_6845384 | 0.11 |
ENST00000303635.7
|
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr6_+_139349903 | 0.11 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr14_+_88471468 | 0.11 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr2_-_17935027 | 0.11 |
ENST00000446852.1
|
SMC6
|
structural maintenance of chromosomes 6 |
| chr22_-_31503490 | 0.11 |
ENST00000400299.2
|
SELM
|
Selenoprotein M |
| chr3_+_156799587 | 0.11 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr19_-_1155118 | 0.11 |
ENST00000590998.1
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr21_+_34602680 | 0.11 |
ENST00000447980.1
|
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr5_-_111093406 | 0.11 |
ENST00000379671.3
|
NREP
|
neuronal regeneration related protein |
| chr19_-_15590306 | 0.11 |
ENST00000292609.4
|
PGLYRP2
|
peptidoglycan recognition protein 2 |
| chr12_+_56618102 | 0.11 |
ENST00000267023.4
ENST00000380198.2 ENST00000341463.5 |
NABP2
|
nucleic acid binding protein 2 |
| chr11_+_73000449 | 0.11 |
ENST00000535931.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr14_-_81408031 | 0.11 |
ENST00000216517.6
|
CEP128
|
centrosomal protein 128kDa |
| chr6_-_18265050 | 0.11 |
ENST00000397239.3
|
DEK
|
DEK oncogene |
| chr22_-_41258074 | 0.11 |
ENST00000307221.4
|
DNAJB7
|
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr5_-_67730240 | 0.11 |
ENST00000507733.1
|
CTC-537E7.3
|
CTC-537E7.3 |
| chr11_-_82997420 | 0.11 |
ENST00000455220.2
ENST00000529689.1 |
CCDC90B
|
coiled-coil domain containing 90B |
| chr2_-_99797473 | 0.11 |
ENST00000409107.1
ENST00000289359.2 |
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr19_-_16606988 | 0.11 |
ENST00000269881.3
|
CALR3
|
calreticulin 3 |
| chr17_-_41132088 | 0.11 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr3_-_160117301 | 0.11 |
ENST00000326448.7
ENST00000498409.1 ENST00000475677.1 ENST00000478536.1 |
IFT80
|
intraflagellar transport 80 homolog (Chlamydomonas) |
| chr17_+_77021702 | 0.11 |
ENST00000392445.2
ENST00000354124.3 |
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr16_-_66907139 | 0.11 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr19_-_55672037 | 0.10 |
ENST00000588076.1
|
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr6_-_110012380 | 0.10 |
ENST00000424296.2
ENST00000341338.6 ENST00000368948.2 ENST00000285397.5 |
AK9
|
adenylate kinase 9 |
| chr11_-_8290263 | 0.10 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr4_+_40194609 | 0.10 |
ENST00000508513.1
|
RHOH
|
ras homolog family member H |
| chr5_-_22853429 | 0.10 |
ENST00000504376.2
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2) |
| chr14_-_81408063 | 0.10 |
ENST00000557411.1
|
CEP128
|
centrosomal protein 128kDa |
| chr14_+_56127960 | 0.10 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr4_+_3344141 | 0.10 |
ENST00000306648.7
|
RGS12
|
regulator of G-protein signaling 12 |
| chr6_-_127840048 | 0.10 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr8_+_7705398 | 0.10 |
ENST00000400125.2
ENST00000434307.2 ENST00000326558.5 ENST00000351436.4 ENST00000528033.1 |
SPAG11A
|
sperm associated antigen 11A |
| chr2_-_172967621 | 0.10 |
ENST00000234198.4
ENST00000466293.2 |
DLX2
|
distal-less homeobox 2 |
| chr18_-_67629015 | 0.10 |
ENST00000579496.1
|
CD226
|
CD226 molecule |
| chr19_-_44405623 | 0.10 |
ENST00000591815.1
|
RP11-15A1.3
|
RP11-15A1.3 |
| chr13_+_74805561 | 0.10 |
ENST00000419499.1
|
LINC00402
|
long intergenic non-protein coding RNA 402 |
| chr8_+_27629459 | 0.10 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr12_+_53443963 | 0.10 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr19_+_10196981 | 0.10 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr1_-_161208013 | 0.10 |
ENST00000515452.1
ENST00000367983.4 |
NR1I3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_+_153631130 | 0.10 |
ENST00000368685.5
|
SNAPIN
|
SNAP-associated protein |
| chr2_-_99797390 | 0.10 |
ENST00000422537.2
|
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr19_-_7167989 | 0.10 |
ENST00000600492.1
|
INSR
|
insulin receptor |
| chr4_-_1657135 | 0.10 |
ENST00000489029.1
|
FAM53A
|
family with sequence similarity 53, member A |
| chr2_-_106013207 | 0.10 |
ENST00000447958.1
|
FHL2
|
four and a half LIM domains 2 |
| chr11_-_82997371 | 0.10 |
ENST00000525503.1
|
CCDC90B
|
coiled-coil domain containing 90B |
| chr14_-_24553834 | 0.10 |
ENST00000397002.2
|
NRL
|
neural retina leucine zipper |
| chr7_+_155755248 | 0.10 |
ENST00000377722.2
|
AC021218.2
|
Uncharacterized protein |
| chr15_+_75335604 | 0.10 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr3_+_118905564 | 0.10 |
ENST00000460625.1
|
UPK1B
|
uroplakin 1B |
| chr19_+_18530184 | 0.10 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr5_+_93954039 | 0.09 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chrX_-_138914394 | 0.09 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr22_-_50970919 | 0.09 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT1_STAT3_BCL6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.2 | 0.7 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.2 | 0.6 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.7 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.1 | 0.4 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.3 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.5 | GO:0072255 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 1.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.2 | GO:0061570 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 0.2 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 2.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0032826 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) |
| 0.0 | 0.2 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.1 | GO:0035712 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.3 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.1 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.2 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.4 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.4 | GO:1900115 | extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.3 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.0 | GO:0061461 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.0 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:0001807 | regulation of type IV hypersensitivity(GO:0001807) |
| 0.0 | 0.1 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.0 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0070092 | glucagon secretion(GO:0070091) regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.0 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 1.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.4 | GO:0042007 | interleukin-18 binding(GO:0042007) |
| 0.1 | 0.3 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.1 | 0.2 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.1 | 0.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.3 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.3 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.1 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.4 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 2.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 1.0 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.5 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |