Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
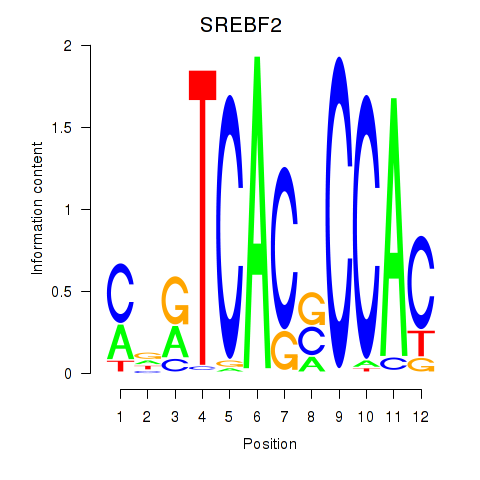
Results for SREBF2
Z-value: 0.52
Transcription factors associated with SREBF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SREBF2
|
ENSG00000198911.7 | sterol regulatory element binding transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SREBF2 | hg19_v2_chr22_+_42229100_42229146 | 0.71 | 1.1e-01 | Click! |
Activity profile of SREBF2 motif
Sorted Z-values of SREBF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_159656437 | 0.48 |
ENST00000402432.3
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr9_+_96928516 | 0.32 |
ENST00000602703.1
|
RP11-2B6.3
|
RP11-2B6.3 |
| chr15_-_73075964 | 0.25 |
ENST00000563907.1
|
ADPGK
|
ADP-dependent glucokinase |
| chr2_-_26205550 | 0.22 |
ENST00000405914.1
|
KIF3C
|
kinesin family member 3C |
| chr13_+_24844857 | 0.22 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr5_-_149669612 | 0.20 |
ENST00000510347.1
|
CAMK2A
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr13_+_24844819 | 0.20 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr17_-_40075197 | 0.18 |
ENST00000590770.1
ENST00000590151.1 |
ACLY
|
ATP citrate lyase |
| chr1_-_42384343 | 0.18 |
ENST00000372584.1
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr9_-_117267717 | 0.18 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr19_+_11200038 | 0.18 |
ENST00000558518.1
ENST00000557933.1 ENST00000455727.2 ENST00000535915.1 ENST00000545707.1 ENST00000558013.1 |
LDLR
|
low density lipoprotein receptor |
| chr6_-_39902160 | 0.18 |
ENST00000340692.5
|
MOCS1
|
molybdenum cofactor synthesis 1 |
| chrX_-_153237258 | 0.17 |
ENST00000310441.7
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr11_+_65480222 | 0.17 |
ENST00000534681.1
|
KAT5
|
K(lysine) acetyltransferase 5 |
| chr2_-_26205340 | 0.17 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chrX_-_153236819 | 0.17 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chrX_-_153236620 | 0.17 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr6_-_31926208 | 0.16 |
ENST00000454913.1
ENST00000436289.2 |
NELFE
|
negative elongation factor complex member E |
| chr6_+_41604747 | 0.16 |
ENST00000419164.1
ENST00000373051.2 |
MDFI
|
MyoD family inhibitor |
| chr1_-_6659876 | 0.16 |
ENST00000496707.1
|
KLHL21
|
kelch-like family member 21 |
| chr19_-_11308190 | 0.16 |
ENST00000586659.1
ENST00000592903.1 ENST00000589359.1 ENST00000588724.1 ENST00000432929.2 |
KANK2
|
KN motif and ankyrin repeat domains 2 |
| chr17_-_40075219 | 0.15 |
ENST00000537919.1
ENST00000352035.2 ENST00000353196.1 ENST00000393896.2 |
ACLY
|
ATP citrate lyase |
| chrX_+_27826107 | 0.15 |
ENST00000356790.2
|
MAGEB10
|
melanoma antigen family B, 10 |
| chr11_+_65479462 | 0.14 |
ENST00000377046.3
ENST00000352980.4 ENST00000341318.4 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr15_-_73076030 | 0.14 |
ENST00000311669.8
|
ADPGK
|
ADP-dependent glucokinase |
| chr16_+_66914264 | 0.14 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr8_+_26434578 | 0.13 |
ENST00000493789.2
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr22_+_42229100 | 0.13 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr20_+_58533471 | 0.12 |
ENST00000244047.5
ENST00000348616.4 |
CDH26
|
cadherin 26 |
| chr18_-_21166841 | 0.12 |
ENST00000269228.5
|
NPC1
|
Niemann-Pick disease, type C1 |
| chr6_+_79577189 | 0.12 |
ENST00000369940.2
|
IRAK1BP1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr1_+_39796810 | 0.12 |
ENST00000289893.4
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr6_+_151561085 | 0.12 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr1_+_55505184 | 0.12 |
ENST00000302118.5
|
PCSK9
|
proprotein convertase subtilisin/kexin type 9 |
| chr17_-_73257667 | 0.12 |
ENST00000538886.1
ENST00000580799.1 ENST00000351904.7 ENST00000537686.1 |
GGA3
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr3_-_16554403 | 0.11 |
ENST00000449415.1
ENST00000441460.1 |
RFTN1
|
raftlin, lipid raft linker 1 |
| chr11_+_65479702 | 0.11 |
ENST00000530446.1
ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr20_+_33464368 | 0.10 |
ENST00000484354.1
ENST00000493805.2 ENST00000473172.1 |
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr5_+_137774706 | 0.10 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr19_-_18392422 | 0.10 |
ENST00000252818.3
|
JUND
|
jun D proto-oncogene |
| chr19_+_36266417 | 0.10 |
ENST00000378944.5
ENST00000007510.4 |
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr5_-_149535421 | 0.10 |
ENST00000261799.4
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide |
| chr11_-_73309228 | 0.09 |
ENST00000356467.4
ENST00000064778.4 |
FAM168A
|
family with sequence similarity 168, member A |
| chr12_-_53343560 | 0.09 |
ENST00000548998.1
|
KRT8
|
keratin 8 |
| chr12_-_53343633 | 0.09 |
ENST00000546826.1
|
KRT8
|
keratin 8 |
| chr11_+_65383227 | 0.09 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr2_+_120517717 | 0.09 |
ENST00000420482.1
ENST00000488279.2 |
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr14_-_74959978 | 0.08 |
ENST00000541064.1
|
NPC2
|
Niemann-Pick disease, type C2 |
| chr1_+_206643787 | 0.08 |
ENST00000367120.3
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr11_+_2923499 | 0.08 |
ENST00000449793.2
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr13_+_24844979 | 0.08 |
ENST00000454083.1
|
SPATA13
|
spermatogenesis associated 13 |
| chr10_-_3827371 | 0.07 |
ENST00000469435.1
|
KLF6
|
Kruppel-like factor 6 |
| chr12_-_53343602 | 0.07 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr17_-_17726907 | 0.07 |
ENST00000423161.3
|
SREBF1
|
sterol regulatory element binding transcription factor 1 |
| chr19_+_38664224 | 0.07 |
ENST00000601054.1
|
SIPA1L3
|
signal-induced proliferation-associated 1 like 3 |
| chr19_-_46285646 | 0.07 |
ENST00000458663.2
|
DMPK
|
dystrophia myotonica-protein kinase |
| chr19_+_36266433 | 0.07 |
ENST00000314737.5
|
ARHGAP33
|
Rho GTPase activating protein 33 |
| chr1_+_33219592 | 0.07 |
ENST00000373481.3
|
KIAA1522
|
KIAA1522 |
| chr16_+_50187556 | 0.07 |
ENST00000561678.1
ENST00000357464.3 |
PAPD5
|
PAP associated domain containing 5 |
| chr19_-_46285736 | 0.07 |
ENST00000291270.4
ENST00000447742.2 ENST00000354227.5 |
DMPK
|
dystrophia myotonica-protein kinase |
| chrX_-_112084043 | 0.06 |
ENST00000304758.1
|
AMOT
|
angiomotin |
| chr3_+_5020801 | 0.06 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr11_+_2923423 | 0.06 |
ENST00000312221.5
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr10_-_3827417 | 0.05 |
ENST00000497571.1
ENST00000542957.1 |
KLF6
|
Kruppel-like factor 6 |
| chrX_+_47229982 | 0.05 |
ENST00000377073.3
|
ZNF157
|
zinc finger protein 157 |
| chr14_-_74960030 | 0.05 |
ENST00000553490.1
ENST00000557510.1 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr14_-_74959994 | 0.04 |
ENST00000238633.2
ENST00000434013.2 |
NPC2
|
Niemann-Pick disease, type C2 |
| chr2_+_232573222 | 0.04 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr19_-_55791058 | 0.04 |
ENST00000587959.1
ENST00000585927.1 ENST00000587922.1 ENST00000585698.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr6_-_32151999 | 0.03 |
ENST00000375069.3
ENST00000538695.1 ENST00000438221.2 ENST00000375065.5 ENST00000450110.1 ENST00000375067.3 ENST00000375056.2 |
AGER
|
advanced glycosylation end product-specific receptor |
| chr5_-_139283982 | 0.03 |
ENST00000340391.3
|
NRG2
|
neuregulin 2 |
| chr20_+_33464238 | 0.03 |
ENST00000360596.2
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr20_+_33464407 | 0.03 |
ENST00000253382.5
|
ACSS2
|
acyl-CoA synthetase short-chain family member 2 |
| chr14_+_74960423 | 0.03 |
ENST00000556816.1
ENST00000298818.8 ENST00000554924.1 |
ISCA2
|
iron-sulfur cluster assembly 2 |
| chr1_+_206643806 | 0.03 |
ENST00000537984.1
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr6_+_43968306 | 0.03 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chrX_-_103087136 | 0.03 |
ENST00000243298.2
|
RAB9B
|
RAB9B, member RAS oncogene family |
| chr2_+_27435734 | 0.03 |
ENST00000419744.1
|
ATRAID
|
all-trans retinoic acid-induced differentiation factor |
| chr8_+_26435359 | 0.03 |
ENST00000311151.5
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr1_-_155232221 | 0.02 |
ENST00000355379.3
|
SCAMP3
|
secretory carrier membrane protein 3 |
| chr2_+_232573208 | 0.02 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr11_+_2923619 | 0.02 |
ENST00000380574.1
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr3_+_46742823 | 0.02 |
ENST00000326431.3
|
TMIE
|
transmembrane inner ear |
| chr10_-_116444371 | 0.02 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr2_+_232572361 | 0.01 |
ENST00000409321.1
|
PTMA
|
prothymosin, alpha |
| chr17_-_15168624 | 0.01 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr11_-_73309112 | 0.01 |
ENST00000450446.2
|
FAM168A
|
family with sequence similarity 168, member A |
| chr17_-_56065484 | 0.01 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr6_+_41604620 | 0.01 |
ENST00000432027.1
|
MDFI
|
MyoD family inhibitor |
| chr17_+_73257742 | 0.01 |
ENST00000579761.1
ENST00000245539.6 |
MRPS7
|
mitochondrial ribosomal protein S7 |
| chr4_+_718896 | 0.00 |
ENST00000433814.1
|
PCGF3
|
polycomb group ring finger 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SREBF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.2 | GO:1903979 | negative regulation of microglial cell activation(GO:1903979) |
| 0.0 | 0.1 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.1 | GO:0060981 | cell migration involved in coronary angiogenesis(GO:0060981) |
| 0.0 | 0.2 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.5 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.2 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.4 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.1 | 0.3 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.1 | 0.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.5 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle binding(GO:0034189) very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.2 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |