Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
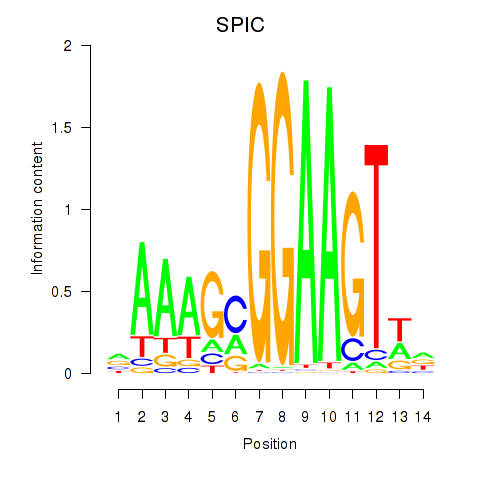
Results for SPIC
Z-value: 0.60
Transcription factors associated with SPIC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIC
|
ENSG00000166211.6 | Spi-C transcription factor |
Activity profile of SPIC motif
Sorted Z-values of SPIC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_39303576 | 0.36 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr1_-_53833812 | 0.28 |
ENST00000449958.1
|
RP11-117D22.2
|
RP11-117D22.2 |
| chr19_+_42381337 | 0.26 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr2_+_138722028 | 0.24 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr19_-_2090131 | 0.20 |
ENST00000591326.1
|
MOB3A
|
MOB kinase activator 3A |
| chr1_+_112016414 | 0.19 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr6_+_139349903 | 0.19 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr16_+_28505955 | 0.19 |
ENST00000564831.1
ENST00000328423.5 ENST00000431282.1 |
APOBR
|
apolipoprotein B receptor |
| chr17_-_4710288 | 0.18 |
ENST00000571067.1
|
RP11-81A22.5
|
RP11-81A22.5 |
| chr7_+_74188309 | 0.17 |
ENST00000289473.4
ENST00000433458.1 |
NCF1
|
neutrophil cytosolic factor 1 |
| chr12_-_54778244 | 0.17 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr4_-_177116772 | 0.17 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr4_-_103682071 | 0.17 |
ENST00000505239.1
|
MANBA
|
mannosidase, beta A, lysosomal |
| chrX_+_47342970 | 0.16 |
ENST00000357412.1
|
CXorf24
|
chromosome X open reading frame 24 |
| chr7_+_95401877 | 0.16 |
ENST00000524053.1
ENST00000324972.6 ENST00000537881.1 ENST00000437599.1 ENST00000359388.4 ENST00000413338.1 |
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr17_+_42925270 | 0.15 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr16_-_4850471 | 0.15 |
ENST00000592019.1
ENST00000586153.1 |
ROGDI
|
rogdi homolog (Drosophila) |
| chr2_+_175352114 | 0.15 |
ENST00000444196.1
ENST00000417038.1 ENST00000606406.1 |
AC010894.3
|
AC010894.3 |
| chr19_+_35940486 | 0.14 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr2_-_64751227 | 0.14 |
ENST00000561559.1
|
RP11-568N6.1
|
RP11-568N6.1 |
| chr17_-_20370847 | 0.14 |
ENST00000423676.3
ENST00000324290.5 |
LGALS9B
|
lectin, galactoside-binding, soluble, 9B |
| chr18_-_61089611 | 0.14 |
ENST00000591519.1
|
VPS4B
|
vacuolar protein sorting 4 homolog B (S. cerevisiae) |
| chr11_+_327171 | 0.13 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr12_+_112451222 | 0.13 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr4_+_123653882 | 0.13 |
ENST00000433287.1
|
BBS12
|
Bardet-Biedl syndrome 12 |
| chr3_+_184530173 | 0.13 |
ENST00000453056.1
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr5_-_138862326 | 0.13 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr4_-_25161996 | 0.13 |
ENST00000513285.1
ENST00000382103.2 |
SEPSECS
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr3_+_156544057 | 0.13 |
ENST00000498839.1
ENST00000470811.1 ENST00000356539.4 ENST00000483177.1 ENST00000477399.1 ENST00000491763.1 |
LEKR1
|
leucine, glutamate and lysine rich 1 |
| chr2_+_217363826 | 0.13 |
ENST00000441179.2
|
RPL37A
|
ribosomal protein L37a |
| chr19_+_35939154 | 0.13 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr7_+_150382781 | 0.13 |
ENST00000223293.5
ENST00000474605.1 |
GIMAP2
|
GTPase, IMAP family member 2 |
| chr11_-_7698453 | 0.12 |
ENST00000524608.1
|
CYB5R2
|
cytochrome b5 reductase 2 |
| chr1_-_183560011 | 0.12 |
ENST00000367536.1
|
NCF2
|
neutrophil cytosolic factor 2 |
| chr1_+_150245177 | 0.12 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr1_+_151009054 | 0.12 |
ENST00000295294.7
|
BNIPL
|
BCL2/adenovirus E1B 19kD interacting protein like |
| chr10_-_1034237 | 0.12 |
ENST00000381466.1
|
AL359878.1
|
Uncharacterized protein |
| chr19_+_35810164 | 0.12 |
ENST00000598537.1
|
CD22
|
CD22 molecule |
| chr7_+_108210012 | 0.12 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr5_-_146461027 | 0.12 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_-_242162375 | 0.12 |
ENST00000357246.3
|
MAP1LC3C
|
microtubule-associated protein 1 light chain 3 gamma |
| chr3_+_57094469 | 0.12 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr17_-_72619869 | 0.12 |
ENST00000392619.1
ENST00000426295.2 |
CD300E
|
CD300e molecule |
| chr9_+_215158 | 0.11 |
ENST00000479404.1
|
DOCK8
|
dedicator of cytokinesis 8 |
| chr6_+_26087646 | 0.11 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr14_-_67826538 | 0.11 |
ENST00000553687.1
|
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr15_-_90233907 | 0.11 |
ENST00000561224.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chrX_-_51239425 | 0.11 |
ENST00000375992.3
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr3_-_142720267 | 0.11 |
ENST00000597953.1
|
RP11-91G21.1
|
RP11-91G21.1 |
| chr11_-_119993979 | 0.11 |
ENST00000524816.3
ENST00000525327.1 |
TRIM29
|
tripartite motif containing 29 |
| chr15_-_33360342 | 0.11 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr6_-_56707943 | 0.11 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr19_+_9732156 | 0.11 |
ENST00000586614.1
ENST00000587536.1 ENST00000591056.1 ENST00000592851.1 |
C19orf82
|
chromosome 19 open reading frame 82 |
| chr7_-_105319536 | 0.11 |
ENST00000477775.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr6_-_170151603 | 0.11 |
ENST00000366774.3
|
TCTE3
|
t-complex-associated-testis-expressed 3 |
| chr5_+_174151536 | 0.10 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr22_-_37880543 | 0.10 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr14_+_88471468 | 0.10 |
ENST00000267549.3
|
GPR65
|
G protein-coupled receptor 65 |
| chr19_+_49838653 | 0.10 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr2_+_63277927 | 0.10 |
ENST00000282549.2
|
OTX1
|
orthodenticle homeobox 1 |
| chr6_+_116782527 | 0.10 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr2_+_217363793 | 0.10 |
ENST00000456586.1
ENST00000598925.1 ENST00000427280.2 |
RPL37A
|
ribosomal protein L37a |
| chr6_+_139349817 | 0.10 |
ENST00000367660.3
|
ABRACL
|
ABRA C-terminal like |
| chr6_-_56708459 | 0.10 |
ENST00000370788.2
|
DST
|
dystonin |
| chr1_+_156698743 | 0.10 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr4_+_123653807 | 0.10 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr10_+_81891416 | 0.10 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr12_+_76653611 | 0.10 |
ENST00000550380.1
|
RP11-54A9.1
|
RP11-54A9.1 |
| chr19_+_58694742 | 0.10 |
ENST00000597528.1
ENST00000594839.1 |
ZNF274
|
zinc finger protein 274 |
| chr2_+_219724544 | 0.10 |
ENST00000233948.3
|
WNT6
|
wingless-type MMTV integration site family, member 6 |
| chr8_+_39770803 | 0.10 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr8_+_86019382 | 0.10 |
ENST00000360375.3
|
LRRCC1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr14_-_89883412 | 0.10 |
ENST00000557258.1
|
FOXN3
|
forkhead box N3 |
| chr8_+_97657531 | 0.10 |
ENST00000519900.1
ENST00000517742.1 |
CPQ
|
carboxypeptidase Q |
| chr15_+_67547113 | 0.10 |
ENST00000512104.1
ENST00000358767.3 ENST00000546225.1 |
IQCH
|
IQ motif containing H |
| chr2_-_264024 | 0.10 |
ENST00000403712.2
ENST00000356150.5 ENST00000405430.1 |
SH3YL1
|
SH3 and SYLF domain containing 1 |
| chr16_+_80574854 | 0.09 |
ENST00000305904.6
ENST00000568035.1 |
DYNLRB2
|
dynein, light chain, roadblock-type 2 |
| chr18_-_11908272 | 0.09 |
ENST00000592977.1
ENST00000590501.1 ENST00000586844.1 |
MPPE1
|
metallophosphoesterase 1 |
| chr19_-_12780211 | 0.09 |
ENST00000597961.1
ENST00000598732.1 ENST00000222190.5 |
CTD-2192J16.24
WDR83OS
|
Uncharacterized protein WD repeat domain 83 opposite strand |
| chr12_-_110888103 | 0.09 |
ENST00000426440.1
ENST00000228825.7 |
ARPC3
|
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr6_+_31553978 | 0.09 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chrX_+_123095860 | 0.09 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr1_+_150245099 | 0.09 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr1_-_9129735 | 0.09 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr1_-_183559693 | 0.09 |
ENST00000367535.3
ENST00000413720.1 ENST00000418089.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr7_-_102252589 | 0.09 |
ENST00000520042.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr1_-_71546690 | 0.09 |
ENST00000254821.6
|
ZRANB2
|
zinc finger, RAN-binding domain containing 2 |
| chr10_-_99161033 | 0.09 |
ENST00000315563.6
ENST00000370992.4 ENST00000414986.1 |
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr11_+_94706804 | 0.09 |
ENST00000335080.5
|
KDM4D
|
lysine (K)-specific demethylase 4D |
| chr2_+_217363559 | 0.09 |
ENST00000600880.1
ENST00000446558.1 |
RPL37A
|
ribosomal protein L37a |
| chr5_-_138861926 | 0.09 |
ENST00000510817.1
|
TMEM173
|
transmembrane protein 173 |
| chr11_-_71639446 | 0.09 |
ENST00000534704.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr19_-_14016877 | 0.09 |
ENST00000454313.1
ENST00000591586.1 ENST00000346736.2 |
C19orf57
|
chromosome 19 open reading frame 57 |
| chr7_-_32529973 | 0.09 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr3_-_145940126 | 0.08 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr17_+_74723031 | 0.08 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr15_+_75335604 | 0.08 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr12_+_51318513 | 0.08 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr7_-_128001658 | 0.08 |
ENST00000489835.2
ENST00000464607.1 ENST00000489517.1 ENST00000446477.2 ENST00000535159.1 ENST00000435512.1 ENST00000495931.1 |
PRRT4
|
proline-rich transmembrane protein 4 |
| chr11_+_112047087 | 0.08 |
ENST00000526088.1
ENST00000532593.1 ENST00000531169.1 |
BCO2
|
beta-carotene oxygenase 2 |
| chr14_-_45722360 | 0.08 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chrX_+_51075658 | 0.08 |
ENST00000356450.2
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr9_-_134145880 | 0.08 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr5_-_114632307 | 0.08 |
ENST00000506442.1
ENST00000379611.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr17_-_74489215 | 0.08 |
ENST00000585701.1
ENST00000591192.1 ENST00000589526.1 |
RHBDF2
|
rhomboid 5 homolog 2 (Drosophila) |
| chr14_-_55738788 | 0.08 |
ENST00000556183.1
|
RP11-665C16.6
|
RP11-665C16.6 |
| chr3_-_47324008 | 0.08 |
ENST00000425853.1
|
KIF9
|
kinesin family member 9 |
| chr15_-_75917955 | 0.08 |
ENST00000568162.1
ENST00000563875.1 |
SNUPN
|
snurportin 1 |
| chr6_-_90121789 | 0.08 |
ENST00000359203.3
|
RRAGD
|
Ras-related GTP binding D |
| chr17_+_7487146 | 0.08 |
ENST00000396501.4
ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1
|
mannose-P-dolichol utilization defect 1 |
| chr2_-_175351744 | 0.08 |
ENST00000295500.4
ENST00000392552.2 ENST00000392551.2 |
GPR155
|
G protein-coupled receptor 155 |
| chr22_-_30942669 | 0.08 |
ENST00000402034.2
|
SEC14L6
|
SEC14-like 6 (S. cerevisiae) |
| chr19_-_9695169 | 0.08 |
ENST00000586602.1
|
ZNF121
|
zinc finger protein 121 |
| chr5_+_115177178 | 0.08 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr1_+_171283331 | 0.07 |
ENST00000367749.3
|
FMO4
|
flavin containing monooxygenase 4 |
| chr5_-_159797627 | 0.07 |
ENST00000393975.3
|
C1QTNF2
|
C1q and tumor necrosis factor related protein 2 |
| chr4_-_164534657 | 0.07 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr6_+_148593425 | 0.07 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr1_+_151009035 | 0.07 |
ENST00000368931.3
|
BNIPL
|
BCL2/adenovirus E1B 19kD interacting protein like |
| chr16_+_25703274 | 0.07 |
ENST00000331351.5
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chr19_+_10222189 | 0.07 |
ENST00000321826.4
|
P2RY11
|
purinergic receptor P2Y, G-protein coupled, 11 |
| chr9_-_7800067 | 0.07 |
ENST00000358227.4
|
TMEM261
|
transmembrane protein 261 |
| chr10_+_22605374 | 0.07 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr12_-_15114603 | 0.07 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr6_-_8435706 | 0.07 |
ENST00000379660.4
|
SLC35B3
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
| chr19_-_5838768 | 0.07 |
ENST00000527106.1
ENST00000531199.1 ENST00000529165.1 |
FUT6
|
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr14_+_74417192 | 0.07 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chrX_-_63615297 | 0.07 |
ENST00000374852.3
ENST00000453546.1 |
MTMR8
|
myotubularin related protein 8 |
| chr9_-_5304432 | 0.07 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr15_-_90234006 | 0.07 |
ENST00000300056.3
ENST00000559170.1 |
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr19_+_1752372 | 0.07 |
ENST00000382349.4
|
ONECUT3
|
one cut homeobox 3 |
| chr7_+_139025875 | 0.07 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr1_+_212968025 | 0.07 |
ENST00000530399.2
|
TATDN3
|
TatD DNase domain containing 3 |
| chr11_+_6226782 | 0.07 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr16_+_57576584 | 0.07 |
ENST00000340339.4
|
GPR114
|
G protein-coupled receptor 114 |
| chr9_+_273038 | 0.07 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr6_+_116601330 | 0.07 |
ENST00000449314.1
ENST00000453463.1 |
RP1-93H18.1
|
RP1-93H18.1 |
| chr4_-_70653673 | 0.07 |
ENST00000512870.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr19_+_56813305 | 0.07 |
ENST00000593151.1
|
AC006116.20
|
Uncharacterized protein |
| chr11_+_1874200 | 0.07 |
ENST00000311604.3
|
LSP1
|
lymphocyte-specific protein 1 |
| chr5_-_114598548 | 0.07 |
ENST00000379615.3
ENST00000419445.1 |
PGGT1B
|
protein geranylgeranyltransferase type I, beta subunit |
| chr16_-_15736881 | 0.07 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr2_+_233925064 | 0.07 |
ENST00000359570.5
ENST00000538935.1 |
INPP5D
|
inositol polyphosphate-5-phosphatase, 145kDa |
| chr4_-_103682145 | 0.07 |
ENST00000226578.4
|
MANBA
|
mannosidase, beta A, lysosomal |
| chr19_+_19626531 | 0.07 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr5_+_68530668 | 0.07 |
ENST00000506563.1
|
CDK7
|
cyclin-dependent kinase 7 |
| chr1_+_145470504 | 0.07 |
ENST00000323397.4
|
ANKRD34A
|
ankyrin repeat domain 34A |
| chr10_+_85954377 | 0.07 |
ENST00000332904.3
ENST00000372117.3 |
CDHR1
|
cadherin-related family member 1 |
| chrY_+_15016725 | 0.07 |
ENST00000336079.3
|
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr3_-_20227720 | 0.07 |
ENST00000412997.1
|
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr7_-_158937544 | 0.07 |
ENST00000421760.2
ENST00000402066.1 |
VIPR2
|
vasoactive intestinal peptide receptor 2 |
| chr11_-_62995986 | 0.07 |
ENST00000403374.2
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr4_-_99578789 | 0.07 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr1_+_33938236 | 0.07 |
ENST00000361328.3
ENST00000373413.2 |
ZSCAN20
|
zinc finger and SCAN domain containing 20 |
| chr5_-_40835303 | 0.07 |
ENST00000509877.1
ENST00000508493.1 ENST00000274242.5 |
RPL37
|
ribosomal protein L37 |
| chr9_+_74526384 | 0.07 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr16_-_66968265 | 0.07 |
ENST00000567511.1
ENST00000422424.2 |
FAM96B
|
family with sequence similarity 96, member B |
| chr20_+_48892848 | 0.07 |
ENST00000422459.1
|
RP11-290F20.3
|
RP11-290F20.3 |
| chr2_+_85822857 | 0.07 |
ENST00000306368.4
ENST00000414390.1 ENST00000456023.1 |
RNF181
|
ring finger protein 181 |
| chr15_-_33360085 | 0.07 |
ENST00000334528.9
|
FMN1
|
formin 1 |
| chrX_-_47489244 | 0.07 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr10_-_119806085 | 0.07 |
ENST00000355624.3
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I) |
| chr9_+_5510492 | 0.07 |
ENST00000397745.2
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr10_+_18948311 | 0.07 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr17_+_18380051 | 0.06 |
ENST00000581545.1
ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C
|
lectin, galactoside-binding, soluble, 9C |
| chr1_+_203830703 | 0.06 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr12_+_12223867 | 0.06 |
ENST00000308721.5
|
BCL2L14
|
BCL2-like 14 (apoptosis facilitator) |
| chrX_-_107018969 | 0.06 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3 |
| chr6_-_110012380 | 0.06 |
ENST00000424296.2
ENST00000341338.6 ENST00000368948.2 ENST00000285397.5 |
AK9
|
adenylate kinase 9 |
| chr4_-_47916543 | 0.06 |
ENST00000507489.1
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr19_+_38826477 | 0.06 |
ENST00000409410.2
ENST00000215069.4 |
CATSPERG
|
catsper channel auxiliary subunit gamma |
| chr15_-_75918500 | 0.06 |
ENST00000569817.1
|
SNUPN
|
snurportin 1 |
| chr11_-_68671244 | 0.06 |
ENST00000567045.1
ENST00000450904.2 |
MRPL21
|
mitochondrial ribosomal protein L21 |
| chr5_-_60458179 | 0.06 |
ENST00000507416.1
ENST00000339020.3 |
SMIM15
|
small integral membrane protein 15 |
| chr19_-_2236290 | 0.06 |
ENST00000591099.2
ENST00000586608.2 ENST00000326631.2 ENST00000587962.2 |
PLEKHJ1
|
pleckstrin homology domain containing, family J member 1 |
| chr15_-_90233866 | 0.06 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr3_-_49142504 | 0.06 |
ENST00000306125.6
ENST00000420147.2 |
QARS
|
glutaminyl-tRNA synthetase |
| chr3_+_10157276 | 0.06 |
ENST00000530758.1
ENST00000256463.6 |
BRK1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr17_+_68047418 | 0.06 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr11_-_32816156 | 0.06 |
ENST00000531481.1
ENST00000335185.5 |
CCDC73
|
coiled-coil domain containing 73 |
| chrX_-_100604184 | 0.06 |
ENST00000372902.3
|
TIMM8A
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr12_-_113841678 | 0.06 |
ENST00000552280.1
ENST00000257549.4 |
SDS
|
serine dehydratase |
| chr19_+_58898627 | 0.06 |
ENST00000598098.1
ENST00000598495.1 ENST00000196551.3 ENST00000596046.1 |
RPS5
|
ribosomal protein S5 |
| chr4_-_99578776 | 0.06 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr4_+_159727272 | 0.06 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr2_-_242211359 | 0.06 |
ENST00000444092.1
|
HDLBP
|
high density lipoprotein binding protein |
| chr5_+_102455853 | 0.06 |
ENST00000515845.1
ENST00000321521.9 ENST00000507921.1 |
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr14_+_32414059 | 0.06 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr17_-_48450534 | 0.06 |
ENST00000503633.1
ENST00000442592.3 ENST00000225969.4 |
MRPL27
|
mitochondrial ribosomal protein L27 |
| chr5_+_6448736 | 0.06 |
ENST00000399816.3
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr8_+_121457642 | 0.06 |
ENST00000305949.1
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr2_+_102758210 | 0.06 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr9_-_34620440 | 0.06 |
ENST00000421919.1
ENST00000378911.3 ENST00000477738.2 ENST00000341694.2 ENST00000259632.7 ENST00000378913.2 ENST00000378916.4 ENST00000447983.2 |
DCTN3
|
dynactin 3 (p22) |
| chr4_-_47916613 | 0.06 |
ENST00000381538.3
ENST00000329043.3 |
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr9_+_214842 | 0.06 |
ENST00000453981.1
ENST00000432829.2 |
DOCK8
|
dedicator of cytokinesis 8 |
| chr5_+_102455968 | 0.06 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr1_-_175161890 | 0.06 |
ENST00000545251.2
ENST00000423313.1 |
KIAA0040
|
KIAA0040 |
| chr18_-_34408802 | 0.06 |
ENST00000590842.1
|
TPGS2
|
tubulin polyglutamylase complex subunit 2 |
| chr5_+_443280 | 0.06 |
ENST00000508022.1
|
EXOC3
|
exocyst complex component 3 |
| chr19_-_13213662 | 0.06 |
ENST00000264824.4
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.1 | 0.2 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.0 | 0.2 | GO:1903722 | negative regulation of exosomal secretion(GO:1903542) regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:0002316 | follicular B cell differentiation(GO:0002316) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.0 | 0.2 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:0061015 | snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.2 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:2001193 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) negative regulation of CD8-positive, alpha-beta T cell activation(GO:2001186) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.1 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.2 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0061570 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.0 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.5 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.0 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.0 | 0.0 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.0 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.0 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.0 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.2 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.0 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.4 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.0 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.2 | GO:0052845 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0030109 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.0 | 0.1 | GO:0004794 | L-threonine ammonia-lyase activity(GO:0004794) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.0 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.0 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.0 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |