Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
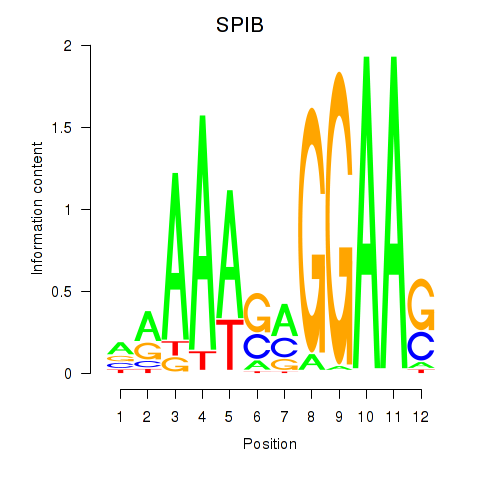
Results for SPIB
Z-value: 0.91
Transcription factors associated with SPIB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIB
|
ENSG00000269404.2 | Spi-B transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIB | hg19_v2_chr19_+_50922187_50922215 | 0.40 | 4.3e-01 | Click! |
Activity profile of SPIB motif
Sorted Z-values of SPIB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_112016414 | 0.76 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chrX_-_47489244 | 0.72 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr15_-_33360342 | 0.59 |
ENST00000558197.1
|
FMN1
|
formin 1 |
| chr19_+_42381337 | 0.42 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr9_+_2017063 | 0.38 |
ENST00000457226.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_-_180397256 | 0.38 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr8_+_121457642 | 0.36 |
ENST00000305949.1
|
MTBP
|
Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa |
| chr8_+_74903580 | 0.36 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr2_-_43019698 | 0.32 |
ENST00000431905.1
ENST00000294973.6 |
HAAO
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr12_-_15114492 | 0.32 |
ENST00000541546.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr6_+_26251835 | 0.31 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr8_+_125860939 | 0.30 |
ENST00000525292.1
ENST00000528090.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr6_-_56708459 | 0.30 |
ENST00000370788.2
|
DST
|
dystonin |
| chr8_-_25281747 | 0.30 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr16_-_4850471 | 0.30 |
ENST00000592019.1
ENST00000586153.1 |
ROGDI
|
rogdi homolog (Drosophila) |
| chr22_-_37880543 | 0.29 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_+_120839412 | 0.29 |
ENST00000355228.4
|
FAM72B
|
family with sequence similarity 72, member B |
| chr15_+_91427691 | 0.29 |
ENST00000559355.1
ENST00000394302.1 |
FES
|
feline sarcoma oncogene |
| chr2_+_138722028 | 0.29 |
ENST00000280096.5
|
HNMT
|
histamine N-methyltransferase |
| chr19_+_35940486 | 0.28 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr3_-_176914191 | 0.28 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chrX_+_24167828 | 0.27 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr15_+_91427642 | 0.27 |
ENST00000328850.3
ENST00000414248.2 |
FES
|
feline sarcoma oncogene |
| chr1_+_173991633 | 0.26 |
ENST00000424181.1
|
RP11-160H22.3
|
RP11-160H22.3 |
| chr1_+_209941827 | 0.26 |
ENST00000367023.1
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr11_-_1606513 | 0.26 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr20_+_20033158 | 0.26 |
ENST00000340348.6
ENST00000377309.2 ENST00000389656.3 ENST00000377306.1 ENST00000245957.5 ENST00000377303.2 ENST00000475466.1 |
C20orf26
|
chromosome 20 open reading frame 26 |
| chr6_-_56707943 | 0.26 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr9_-_95640218 | 0.26 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr15_-_65809581 | 0.25 |
ENST00000341861.5
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr3_+_184530173 | 0.25 |
ENST00000453056.1
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr1_-_45956868 | 0.24 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr17_+_32597232 | 0.24 |
ENST00000378569.2
ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7
|
chemokine (C-C motif) ligand 7 |
| chr8_+_27632083 | 0.24 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr5_-_158757895 | 0.23 |
ENST00000231228.2
|
IL12B
|
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr15_-_33360085 | 0.23 |
ENST00000334528.9
|
FMN1
|
formin 1 |
| chr17_-_27467418 | 0.23 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr3_+_16926441 | 0.23 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr11_-_64512273 | 0.23 |
ENST00000377497.3
ENST00000377487.1 ENST00000430645.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr2_+_196440692 | 0.23 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr8_+_125985531 | 0.22 |
ENST00000319286.5
|
ZNF572
|
zinc finger protein 572 |
| chr19_+_35939154 | 0.21 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chrX_+_123094672 | 0.21 |
ENST00000354548.5
ENST00000458700.1 |
STAG2
|
stromal antigen 2 |
| chr5_-_10249990 | 0.21 |
ENST00000511437.1
ENST00000280330.8 ENST00000510047.1 |
FAM173B
|
family with sequence similarity 173, member B |
| chrX_+_123094369 | 0.21 |
ENST00000455404.1
ENST00000218089.9 |
STAG2
|
stromal antigen 2 |
| chr21_-_32716556 | 0.20 |
ENST00000455508.1
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1 |
| chr15_-_65809625 | 0.20 |
ENST00000560436.1
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr8_-_74206133 | 0.20 |
ENST00000352983.2
|
RPL7
|
ribosomal protein L7 |
| chr3_-_143567262 | 0.20 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr2_-_188419200 | 0.20 |
ENST00000233156.3
ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr2_+_102758210 | 0.20 |
ENST00000450319.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr17_+_65374075 | 0.19 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr7_+_23637118 | 0.19 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr7_-_150038704 | 0.19 |
ENST00000466675.1
ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr12_-_53901266 | 0.19 |
ENST00000609999.1
ENST00000267017.3 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr8_-_121457332 | 0.19 |
ENST00000518918.1
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr4_-_146859787 | 0.19 |
ENST00000508784.1
|
ZNF827
|
zinc finger protein 827 |
| chr9_-_14693417 | 0.18 |
ENST00000380916.4
|
ZDHHC21
|
zinc finger, DHHC-type containing 21 |
| chr1_-_222886526 | 0.18 |
ENST00000541237.1
|
AIDA
|
axin interactor, dorsalization associated |
| chr3_-_114035026 | 0.18 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chrX_+_24167746 | 0.18 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr14_+_102228123 | 0.18 |
ENST00000422945.2
ENST00000554442.1 ENST00000556260.2 ENST00000328724.5 ENST00000557268.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr2_-_64751227 | 0.18 |
ENST00000561559.1
|
RP11-568N6.1
|
RP11-568N6.1 |
| chr8_-_121457608 | 0.18 |
ENST00000306185.3
|
MRPL13
|
mitochondrial ribosomal protein L13 |
| chr19_+_10959043 | 0.17 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr14_+_96968802 | 0.17 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr5_+_102455968 | 0.17 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr12_+_50355647 | 0.17 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr1_+_90287480 | 0.17 |
ENST00000394593.3
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr22_+_17082732 | 0.17 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr13_-_95364389 | 0.17 |
ENST00000376945.2
|
SOX21
|
SRY (sex determining region Y)-box 21 |
| chr14_+_96968707 | 0.17 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr4_-_47916543 | 0.17 |
ENST00000507489.1
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr3_+_23847394 | 0.16 |
ENST00000306627.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr20_-_20033052 | 0.16 |
ENST00000536226.1
|
CRNKL1
|
crooked neck pre-mRNA splicing factor 1 |
| chr1_-_179851611 | 0.16 |
ENST00000610272.1
|
RP11-533E19.7
|
RP11-533E19.7 |
| chr4_+_190861941 | 0.16 |
ENST00000226798.4
|
FRG1
|
FSHD region gene 1 |
| chr6_+_31553978 | 0.16 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr8_-_74205851 | 0.16 |
ENST00000396467.1
|
RPL7
|
ribosomal protein L7 |
| chr8_-_6420759 | 0.16 |
ENST00000523120.1
|
ANGPT2
|
angiopoietin 2 |
| chr2_-_74405929 | 0.16 |
ENST00000396049.4
|
MOB1A
|
MOB kinase activator 1A |
| chr10_-_14596140 | 0.15 |
ENST00000496330.1
|
FAM107B
|
family with sequence similarity 107, member B |
| chr2_-_188419078 | 0.15 |
ENST00000437725.1
ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI
|
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr3_+_23847432 | 0.15 |
ENST00000346855.3
|
UBE2E1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr10_+_114135952 | 0.15 |
ENST00000356116.1
ENST00000433418.1 ENST00000354273.4 |
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr9_+_104296163 | 0.15 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr15_-_83224682 | 0.15 |
ENST00000562833.1
|
RP11-152F13.10
|
RP11-152F13.10 |
| chr6_+_31554826 | 0.15 |
ENST00000376089.2
ENST00000396112.2 |
LST1
|
leukocyte specific transcript 1 |
| chr12_-_15114603 | 0.15 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_+_167298281 | 0.15 |
ENST00000367862.5
|
POU2F1
|
POU class 2 homeobox 1 |
| chr17_-_29641104 | 0.15 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr11_-_8680383 | 0.15 |
ENST00000299550.6
|
TRIM66
|
tripartite motif containing 66 |
| chr15_+_86098670 | 0.15 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr4_+_190861993 | 0.15 |
ENST00000524583.1
ENST00000531991.2 |
FRG1
|
FSHD region gene 1 |
| chr1_+_104068312 | 0.15 |
ENST00000524631.1
ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr4_-_140005341 | 0.14 |
ENST00000379549.2
ENST00000512627.1 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr4_-_47916613 | 0.14 |
ENST00000381538.3
ENST00000329043.3 |
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr14_+_32546145 | 0.14 |
ENST00000556611.1
ENST00000539826.2 |
ARHGAP5
|
Rho GTPase activating protein 5 |
| chr14_+_74417192 | 0.14 |
ENST00000554320.1
|
COQ6
|
coenzyme Q6 monooxygenase |
| chr1_+_66796401 | 0.14 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr19_+_49838653 | 0.14 |
ENST00000598095.1
ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37
|
CD37 molecule |
| chr5_-_64858944 | 0.14 |
ENST00000508421.1
ENST00000510693.1 ENST00000514814.1 ENST00000515497.1 ENST00000396679.1 |
CENPK
|
centromere protein K |
| chr5_-_58335281 | 0.14 |
ENST00000358923.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr8_-_95449155 | 0.14 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr1_-_45956822 | 0.14 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr7_+_86781778 | 0.14 |
ENST00000432937.2
|
DMTF1
|
cyclin D binding myb-like transcription factor 1 |
| chr3_-_16554881 | 0.14 |
ENST00000451036.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr10_-_74856608 | 0.14 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr19_-_48867171 | 0.14 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr13_+_108922228 | 0.13 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr14_-_80678512 | 0.13 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr7_-_37488777 | 0.13 |
ENST00000445322.1
ENST00000448602.1 |
ELMO1
|
engulfment and cell motility 1 |
| chr16_+_31213206 | 0.13 |
ENST00000561916.2
|
C16orf98
|
chromosome 16 open reading frame 98 |
| chr15_+_91427726 | 0.13 |
ENST00000452243.1
|
FES
|
feline sarcoma oncogene |
| chr10_-_65028817 | 0.13 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr4_-_10686475 | 0.13 |
ENST00000226951.6
|
CLNK
|
cytokine-dependent hematopoietic cell linker |
| chr2_+_64751433 | 0.13 |
ENST00000238856.4
ENST00000422803.1 ENST00000238855.7 |
AFTPH
|
aftiphilin |
| chr3_+_239652 | 0.13 |
ENST00000435603.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr14_-_67826486 | 0.13 |
ENST00000555431.1
ENST00000554236.1 ENST00000555474.1 |
ATP6V1D
|
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr15_-_65810042 | 0.13 |
ENST00000321147.6
|
DPP8
|
dipeptidyl-peptidase 8 |
| chr1_+_222886694 | 0.13 |
ENST00000426638.1
ENST00000537020.1 ENST00000539697.1 |
BROX
|
BRO1 domain and CAAX motif containing |
| chr4_-_103266626 | 0.12 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr5_+_118668846 | 0.12 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr3_+_4535155 | 0.12 |
ENST00000544951.1
|
ITPR1
|
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr20_+_5931275 | 0.12 |
ENST00000378896.3
ENST00000378883.1 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr14_+_45431379 | 0.12 |
ENST00000361577.3
ENST00000361462.2 ENST00000382233.2 |
FAM179B
|
family with sequence similarity 179, member B |
| chr1_+_205473720 | 0.12 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr20_+_5931497 | 0.12 |
ENST00000378886.2
ENST00000265187.4 |
MCM8
|
minichromosome maintenance complex component 8 |
| chr7_-_37382360 | 0.12 |
ENST00000455119.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr11_-_47400032 | 0.12 |
ENST00000533968.1
|
SPI1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr6_-_49430886 | 0.12 |
ENST00000274813.3
|
MUT
|
methylmalonyl CoA mutase |
| chr5_+_64859066 | 0.12 |
ENST00000261308.5
ENST00000535264.1 ENST00000538977.1 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr8_-_125551278 | 0.12 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr6_+_142468383 | 0.12 |
ENST00000367621.1
ENST00000452973.2 |
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr5_-_66492562 | 0.12 |
ENST00000256447.4
|
CD180
|
CD180 molecule |
| chr17_+_28443799 | 0.12 |
ENST00000584423.1
ENST00000247026.5 |
NSRP1
|
nuclear speckle splicing regulatory protein 1 |
| chr12_-_122240792 | 0.12 |
ENST00000545885.1
ENST00000542933.1 ENST00000428029.2 ENST00000541694.1 ENST00000536662.1 ENST00000535643.1 ENST00000541657.1 |
AC084018.1
RHOF
|
AC084018.1 ras homolog family member F (in filopodia) |
| chr15_-_66084621 | 0.11 |
ENST00000564674.1
|
DENND4A
|
DENN/MADD domain containing 4A |
| chr20_-_5931051 | 0.11 |
ENST00000453074.2
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr7_-_19157248 | 0.11 |
ENST00000242261.5
|
TWIST1
|
twist family bHLH transcription factor 1 |
| chr1_-_54879140 | 0.11 |
ENST00000525990.1
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr13_+_36050881 | 0.11 |
ENST00000537702.1
|
NBEA
|
neurobeachin |
| chr1_-_43282906 | 0.11 |
ENST00000372521.4
|
CCDC23
|
coiled-coil domain containing 23 |
| chr17_+_7461781 | 0.11 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr9_-_34397800 | 0.11 |
ENST00000297623.2
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr11_-_10920838 | 0.11 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr17_-_72619869 | 0.11 |
ENST00000392619.1
ENST00000426295.2 |
CD300E
|
CD300e molecule |
| chr1_-_17380630 | 0.11 |
ENST00000375499.3
|
SDHB
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr17_+_7462031 | 0.11 |
ENST00000380535.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr5_-_140013241 | 0.11 |
ENST00000519715.1
|
CD14
|
CD14 molecule |
| chr10_+_7830125 | 0.11 |
ENST00000335698.4
ENST00000541227.1 |
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr2_+_102758271 | 0.11 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr11_-_64512803 | 0.11 |
ENST00000377489.1
ENST00000354024.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr6_-_139308777 | 0.11 |
ENST00000529597.1
ENST00000415951.2 ENST00000367663.4 ENST00000409812.2 |
REPS1
|
RALBP1 associated Eps domain containing 1 |
| chr6_+_49431073 | 0.11 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr5_-_39270725 | 0.11 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr2_-_180871780 | 0.11 |
ENST00000410053.3
ENST00000295749.6 ENST00000404136.2 |
CWC22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr1_+_151129103 | 0.11 |
ENST00000368910.3
|
TNFAIP8L2
|
tumor necrosis factor, alpha-induced protein 8-like 2 |
| chr5_-_140013224 | 0.10 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr17_+_7461613 | 0.10 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr6_-_110012380 | 0.10 |
ENST00000424296.2
ENST00000341338.6 ENST00000368948.2 ENST00000285397.5 |
AK9
|
adenylate kinase 9 |
| chr9_+_215158 | 0.10 |
ENST00000479404.1
|
DOCK8
|
dedicator of cytokinesis 8 |
| chr12_+_32638897 | 0.10 |
ENST00000531134.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr7_+_23636992 | 0.10 |
ENST00000307471.3
ENST00000409765.1 |
CCDC126
|
coiled-coil domain containing 126 |
| chr19_-_15344243 | 0.10 |
ENST00000602233.1
|
EPHX3
|
epoxide hydrolase 3 |
| chr6_+_22221010 | 0.10 |
ENST00000567753.1
|
RP11-524C21.2
|
RP11-524C21.2 |
| chr17_-_4642429 | 0.10 |
ENST00000573123.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr11_-_64570624 | 0.10 |
ENST00000439069.1
|
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr6_-_47009996 | 0.10 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr3_-_155462808 | 0.10 |
ENST00000460012.1
|
PLCH1
|
phospholipase C, eta 1 |
| chr9_+_104296243 | 0.10 |
ENST00000466817.1
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr8_-_131028869 | 0.10 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr22_+_20905269 | 0.10 |
ENST00000457322.1
|
MED15
|
mediator complex subunit 15 |
| chr10_-_7829909 | 0.10 |
ENST00000379562.4
ENST00000543003.1 ENST00000535925.1 |
KIN
|
KIN, antigenic determinant of recA protein homolog (mouse) |
| chr3_-_178984759 | 0.10 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr14_+_102276209 | 0.10 |
ENST00000445439.3
ENST00000334743.5 ENST00000557095.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr3_-_176914238 | 0.10 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr1_-_175161890 | 0.10 |
ENST00000545251.2
ENST00000423313.1 |
KIAA0040
|
KIAA0040 |
| chr18_-_32924372 | 0.10 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr2_-_174830430 | 0.10 |
ENST00000310015.6
ENST00000455789.2 |
SP3
|
Sp3 transcription factor |
| chr7_-_135662056 | 0.10 |
ENST00000393085.3
ENST00000435723.1 |
MTPN
|
myotrophin |
| chr12_-_110939870 | 0.10 |
ENST00000447578.2
ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr10_+_7830092 | 0.09 |
ENST00000356708.7
|
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr1_-_31230650 | 0.09 |
ENST00000294507.3
|
LAPTM5
|
lysosomal protein transmembrane 5 |
| chr3_+_184529948 | 0.09 |
ENST00000436792.2
ENST00000446204.2 ENST00000422105.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chrX_+_11777671 | 0.09 |
ENST00000380693.3
ENST00000380692.2 |
MSL3
|
male-specific lethal 3 homolog (Drosophila) |
| chr8_-_77912431 | 0.09 |
ENST00000357039.4
ENST00000522527.1 |
PEX2
|
peroxisomal biogenesis factor 2 |
| chr19_+_51728316 | 0.09 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr8_+_104426942 | 0.09 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr14_+_102276132 | 0.09 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_161551101 | 0.09 |
ENST00000367962.4
ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B
|
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr15_+_50716576 | 0.09 |
ENST00000560297.1
ENST00000307179.4 ENST00000396444.3 ENST00000433963.1 ENST00000425032.3 |
USP8
|
ubiquitin specific peptidase 8 |
| chr15_+_77287715 | 0.09 |
ENST00000559161.1
|
PSTPIP1
|
proline-serine-threonine phosphatase interacting protein 1 |
| chr3_+_184529929 | 0.09 |
ENST00000287546.4
ENST00000437079.3 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr7_+_139529085 | 0.09 |
ENST00000539806.1
|
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr19_-_3479086 | 0.09 |
ENST00000587847.1
|
C19orf77
|
chromosome 19 open reading frame 77 |
| chrX_+_24072833 | 0.09 |
ENST00000253039.4
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr3_-_179169330 | 0.09 |
ENST00000232564.3
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr8_+_27632047 | 0.09 |
ENST00000397418.2
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr16_-_19725899 | 0.09 |
ENST00000567367.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.1 | 0.8 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.3 | GO:1990637 | response to prolactin(GO:1990637) regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.7 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.5 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.1 | 0.3 | GO:0044026 | DNA hypermethylation(GO:0044026) hypermethylation of CpG island(GO:0044027) |
| 0.1 | 0.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.2 | GO:0019732 | antifungal humoral response(GO:0019732) |
| 0.1 | 0.3 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.4 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 | 0.2 | GO:0061570 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 0.1 | GO:0002605 | negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.3 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.2 | GO:2000330 | positive regulation of T-helper 17 cell lineage commitment(GO:2000330) |
| 0.0 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.2 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.3 | GO:0071727 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.0 | 0.4 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.3 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.8 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.2 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0061485 | memory T cell proliferation(GO:0061485) |
| 0.0 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.1 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.0 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0043375 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.3 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.0 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0052056 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0071226 | response to molecule of fungal origin(GO:0002238) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.1 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.0 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.1 | GO:1904879 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.0 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.0 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.0 | 0.1 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.5 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.3 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.3 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.3 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.3 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |