Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
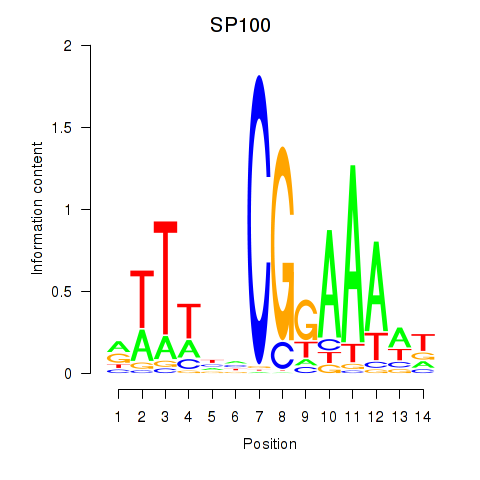
Results for SP100
Z-value: 1.87
Transcription factors associated with SP100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SP100
|
ENSG00000067066.12 | SP100 nuclear antigen |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SP100 | hg19_v2_chr2_+_231280954_231280981 | 0.73 | 9.8e-02 | Click! |
Activity profile of SP100 motif
Sorted Z-values of SP100 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_89299994 | 2.11 |
ENST00000264346.7
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr11_-_615942 | 1.89 |
ENST00000397562.3
ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7
|
interferon regulatory factor 7 |
| chr2_+_114163945 | 1.82 |
ENST00000453673.3
|
IGKV1OR2-108
|
immunoglobulin kappa variable 1/OR2-108 (non-functional) |
| chr7_-_81635106 | 1.65 |
ENST00000443883.1
|
CACNA2D1
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| chr1_-_63988846 | 1.53 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr6_-_86303833 | 1.38 |
ENST00000505648.1
|
SNX14
|
sorting nexin 14 |
| chr6_-_18249971 | 1.37 |
ENST00000507591.1
|
DEK
|
DEK oncogene |
| chr9_-_32526299 | 1.34 |
ENST00000379882.1
ENST00000379883.2 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr9_-_32526184 | 1.32 |
ENST00000545044.1
ENST00000379868.1 |
DDX58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr10_-_112255945 | 1.29 |
ENST00000609514.1
ENST00000607952.1 |
RP11-525A16.4
|
RP11-525A16.4 |
| chr6_-_86303523 | 1.18 |
ENST00000513865.1
ENST00000369627.2 ENST00000514419.1 ENST00000509338.1 ENST00000314673.3 ENST00000346348.3 |
SNX14
|
sorting nexin 14 |
| chr1_+_63989004 | 1.12 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr13_+_114462193 | 1.11 |
ENST00000375353.3
|
TMEM255B
|
transmembrane protein 255B |
| chr5_-_102455801 | 1.05 |
ENST00000508629.1
ENST00000399004.2 |
GIN1
|
gypsy retrotransposon integrase 1 |
| chr4_+_184427235 | 1.02 |
ENST00000412117.1
ENST00000434682.2 |
ING2
|
inhibitor of growth family, member 2 |
| chr2_-_231084659 | 1.02 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr4_+_89299885 | 1.02 |
ENST00000380265.5
ENST00000273960.3 |
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr9_+_79792410 | 0.98 |
ENST00000357409.5
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr1_+_111682058 | 0.95 |
ENST00000545121.1
|
CEPT1
|
choline/ethanolamine phosphotransferase 1 |
| chr1_+_214776516 | 0.94 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr15_+_66797455 | 0.94 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr8_+_94767072 | 0.91 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr10_-_35379524 | 0.90 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chr8_+_27629459 | 0.90 |
ENST00000523566.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr10_+_91174314 | 0.88 |
ENST00000371795.4
|
IFIT5
|
interferon-induced protein with tetratricopeptide repeats 5 |
| chr15_+_66797627 | 0.86 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chrX_-_77395186 | 0.86 |
ENST00000341864.5
|
TAF9B
|
TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa |
| chr14_+_45553296 | 0.85 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr7_-_124569991 | 0.85 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr12_+_95867727 | 0.85 |
ENST00000323666.5
ENST00000546753.1 |
METAP2
|
methionyl aminopeptidase 2 |
| chr4_-_170679024 | 0.85 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
| chr5_+_72143988 | 0.84 |
ENST00000506351.2
|
TNPO1
|
transportin 1 |
| chr14_+_57735725 | 0.84 |
ENST00000431972.2
|
AP5M1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr7_+_131012605 | 0.84 |
ENST00000446815.1
ENST00000352689.6 |
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr5_-_55008072 | 0.83 |
ENST00000512208.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr6_+_96969672 | 0.82 |
ENST00000369278.4
|
UFL1
|
UFM1-specific ligase 1 |
| chr12_-_120884175 | 0.82 |
ENST00000546954.1
|
TRIAP1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr21_-_34863998 | 0.82 |
ENST00000402202.1
ENST00000381947.3 |
DNAJC28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr2_-_231084820 | 0.81 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr5_-_126409159 | 0.80 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr1_+_196621002 | 0.79 |
ENST00000367429.4
ENST00000439155.2 |
CFH
|
complement factor H |
| chr7_+_117824210 | 0.79 |
ENST00000422760.1
ENST00000411938.1 |
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr19_+_10197463 | 0.78 |
ENST00000590378.1
ENST00000397881.3 |
C19orf66
|
chromosome 19 open reading frame 66 |
| chr18_-_32870148 | 0.78 |
ENST00000589178.1
ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30
RP11-158H5.7
|
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chr5_-_87516448 | 0.77 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr2_-_47403642 | 0.77 |
ENST00000456319.1
ENST00000409563.1 ENST00000272298.7 |
CALM2
|
calmodulin 2 (phosphorylase kinase, delta) |
| chr17_-_64216748 | 0.77 |
ENST00000585162.1
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr4_+_25378826 | 0.77 |
ENST00000315368.3
|
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr12_+_49961990 | 0.76 |
ENST00000551063.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr5_+_40841410 | 0.76 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr8_+_90914073 | 0.76 |
ENST00000297438.2
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr1_-_94344754 | 0.76 |
ENST00000436063.2
|
DNTTIP2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr12_+_27863706 | 0.76 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr2_-_223520770 | 0.75 |
ENST00000536361.1
|
FARSB
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr6_+_144164455 | 0.75 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr7_-_35013217 | 0.74 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr12_+_97306295 | 0.73 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr1_+_90098606 | 0.73 |
ENST00000370454.4
|
LRRC8C
|
leucine rich repeat containing 8 family, member C |
| chr6_+_29691198 | 0.71 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr14_-_102605983 | 0.71 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr15_-_67546963 | 0.69 |
ENST00000561452.1
ENST00000261880.5 |
AAGAB
|
alpha- and gamma-adaptin binding protein |
| chr4_+_41937131 | 0.68 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chrM_+_4431 | 0.68 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr6_-_28554977 | 0.66 |
ENST00000452236.2
|
SCAND3
|
SCAN domain containing 3 |
| chr3_+_62304648 | 0.66 |
ENST00000462069.1
ENST00000232519.5 ENST00000465142.1 |
C3orf14
|
chromosome 3 open reading frame 14 |
| chr14_-_54908043 | 0.66 |
ENST00000556113.1
ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr10_+_35416223 | 0.65 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr15_+_72410629 | 0.65 |
ENST00000340912.4
ENST00000544171.1 |
SENP8
|
SUMO/sentrin specific peptidase family member 8 |
| chr10_-_96122682 | 0.65 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr21_-_30365136 | 0.65 |
ENST00000361371.5
ENST00000389194.2 ENST00000389195.2 |
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr10_+_112327425 | 0.65 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr19_-_23433144 | 0.64 |
ENST00000418100.1
ENST00000597537.1 ENST00000597037.1 |
ZNF724P
|
zinc finger protein 724, pseudogene |
| chr7_+_99195878 | 0.64 |
ENST00000453227.1
ENST00000429679.1 |
GS1-259H13.2
|
GS1-259H13.2 |
| chr12_+_69004805 | 0.64 |
ENST00000541216.1
|
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr2_+_201390843 | 0.64 |
ENST00000357799.4
ENST00000409203.3 |
SGOL2
|
shugoshin-like 2 (S. pombe) |
| chr13_-_33112899 | 0.63 |
ENST00000267068.3
ENST00000357505.6 ENST00000399396.3 |
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr9_+_33265011 | 0.63 |
ENST00000419016.2
|
CHMP5
|
charged multivesicular body protein 5 |
| chr16_-_66907139 | 0.63 |
ENST00000561579.2
|
NAE1
|
NEDD8 activating enzyme E1 subunit 1 |
| chr7_-_91764108 | 0.63 |
ENST00000450723.1
|
CYP51A1
|
cytochrome P450, family 51, subfamily A, polypeptide 1 |
| chr6_-_127664475 | 0.63 |
ENST00000474289.2
ENST00000534442.1 ENST00000368289.2 ENST00000525745.1 ENST00000430841.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr14_+_75745477 | 0.62 |
ENST00000303562.4
ENST00000554617.1 ENST00000554212.1 ENST00000535987.1 ENST00000555242.1 |
FOS
|
FBJ murine osteosarcoma viral oncogene homolog |
| chr19_+_33072373 | 0.62 |
ENST00000586035.1
|
PDCD5
|
programmed cell death 5 |
| chr5_-_159846066 | 0.61 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr2_-_37068530 | 0.61 |
ENST00000593798.1
|
AC007382.1
|
Uncharacterized protein |
| chr15_-_55488817 | 0.61 |
ENST00000569386.1
|
RSL24D1
|
ribosomal L24 domain containing 1 |
| chr7_-_124569864 | 0.60 |
ENST00000609702.1
|
POT1
|
protection of telomeres 1 |
| chr2_+_28974489 | 0.60 |
ENST00000455580.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr13_-_31736132 | 0.60 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr8_+_110552337 | 0.60 |
ENST00000337573.5
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr13_-_33112956 | 0.59 |
ENST00000505213.1
|
N4BP2L2
|
NEDD4 binding protein 2-like 2 |
| chr6_+_29691056 | 0.59 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr11_-_93474645 | 0.59 |
ENST00000532455.1
|
TAF1D
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa |
| chr1_-_235324772 | 0.59 |
ENST00000408888.3
|
RBM34
|
RNA binding motif protein 34 |
| chr21_+_34602680 | 0.58 |
ENST00000447980.1
|
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr2_+_157291953 | 0.58 |
ENST00000310454.6
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr14_+_45605127 | 0.58 |
ENST00000556036.1
ENST00000267430.5 |
FANCM
|
Fanconi anemia, complementation group M |
| chr10_-_30637906 | 0.58 |
ENST00000417581.1
|
MTPAP
|
mitochondrial poly(A) polymerase |
| chr11_-_17410869 | 0.58 |
ENST00000528731.1
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr6_+_163837347 | 0.58 |
ENST00000544436.1
|
QKI
|
QKI, KH domain containing, RNA binding |
| chrX_+_13752832 | 0.57 |
ENST00000380550.3
ENST00000398395.3 ENST00000340096.6 ENST00000380567.1 |
OFD1
|
oral-facial-digital syndrome 1 |
| chrX_-_99891796 | 0.57 |
ENST00000373020.4
|
TSPAN6
|
tetraspanin 6 |
| chr1_-_87380002 | 0.57 |
ENST00000331835.5
|
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr22_-_28316116 | 0.56 |
ENST00000415296.1
|
PITPNB
|
phosphatidylinositol transfer protein, beta |
| chr14_-_88200641 | 0.56 |
ENST00000556168.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr3_+_122296465 | 0.56 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr13_+_27825706 | 0.56 |
ENST00000272274.4
ENST00000319826.4 ENST00000326092.4 |
RPL21
|
ribosomal protein L21 |
| chr5_-_38845812 | 0.56 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr2_-_178417742 | 0.56 |
ENST00000408939.3
|
TTC30B
|
tetratricopeptide repeat domain 30B |
| chr14_+_74318611 | 0.56 |
ENST00000555976.1
ENST00000267568.4 |
PTGR2
|
prostaglandin reductase 2 |
| chr8_-_95487331 | 0.56 |
ENST00000336148.5
|
RAD54B
|
RAD54 homolog B (S. cerevisiae) |
| chr6_-_107230334 | 0.56 |
ENST00000607090.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr14_-_63974941 | 0.56 |
ENST00000422769.2
|
PPP2R5E
|
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr2_-_113191096 | 0.55 |
ENST00000496537.1
ENST00000330575.5 ENST00000302558.3 |
RGPD8
|
RANBP2-like and GRIP domain containing 8 |
| chr5_+_32585605 | 0.55 |
ENST00000265073.4
ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1
|
SUB1 homolog (S. cerevisiae) |
| chr10_+_37414785 | 0.55 |
ENST00000374660.1
ENST00000602533.1 ENST00000361713.1 |
ANKRD30A
|
ankyrin repeat domain 30A |
| chr4_+_25378912 | 0.55 |
ENST00000510092.1
ENST00000505991.1 |
ANAPC4
|
anaphase promoting complex subunit 4 |
| chr11_+_18477369 | 0.55 |
ENST00000396213.3
ENST00000280706.2 |
LDHAL6A
|
lactate dehydrogenase A-like 6A |
| chr14_+_45605157 | 0.55 |
ENST00000542564.2
|
FANCM
|
Fanconi anemia, complementation group M |
| chr12_+_97300995 | 0.55 |
ENST00000266742.4
ENST00000429527.2 ENST00000554226.1 ENST00000557478.1 ENST00000557092.1 ENST00000411739.2 ENST00000553609.1 |
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr5_+_179135246 | 0.55 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr7_-_32529973 | 0.54 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr5_+_121297650 | 0.54 |
ENST00000339397.4
|
SRFBP1
|
serum response factor binding protein 1 |
| chr1_+_227751231 | 0.54 |
ENST00000343776.5
ENST00000608949.1 ENST00000397097.3 |
ZNF678
|
zinc finger protein 678 |
| chr14_+_58765103 | 0.53 |
ENST00000355431.3
ENST00000348476.3 ENST00000395168.3 |
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr2_-_99797390 | 0.53 |
ENST00000422537.2
|
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr3_+_62304712 | 0.52 |
ENST00000494481.1
|
C3orf14
|
chromosome 3 open reading frame 14 |
| chr13_+_27825446 | 0.52 |
ENST00000311549.6
|
RPL21
|
ribosomal protein L21 |
| chr7_-_23145288 | 0.52 |
ENST00000419813.1
|
KLHL7-AS1
|
KLHL7 antisense RNA 1 (head to head) |
| chr19_+_53836985 | 0.51 |
ENST00000601857.1
ENST00000595091.1 ENST00000458035.1 |
ZNF845
|
zinc finger protein 845 |
| chr1_+_70876926 | 0.51 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr3_-_108308241 | 0.51 |
ENST00000295746.8
|
KIAA1524
|
KIAA1524 |
| chr10_-_38146482 | 0.51 |
ENST00000374648.3
|
ZNF248
|
zinc finger protein 248 |
| chr12_+_104324112 | 0.51 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr1_-_87379785 | 0.51 |
ENST00000401030.3
ENST00000370554.1 |
SEP15
|
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr11_+_102980126 | 0.51 |
ENST00000375735.2
|
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1 |
| chr17_+_57642886 | 0.51 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr1_+_179335101 | 0.51 |
ENST00000508285.1
ENST00000511889.1 |
AXDND1
|
axonemal dynein light chain domain containing 1 |
| chr5_-_68665296 | 0.51 |
ENST00000512152.1
ENST00000503245.1 ENST00000512561.1 ENST00000380822.4 |
TAF9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr10_+_45495898 | 0.51 |
ENST00000298299.3
|
ZNF22
|
zinc finger protein 22 |
| chr14_+_50065376 | 0.50 |
ENST00000298288.6
|
LRR1
|
leucine rich repeat protein 1 |
| chr1_+_75198871 | 0.50 |
ENST00000479111.1
|
TYW3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr15_+_55611128 | 0.50 |
ENST00000164305.5
ENST00000539642.1 |
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr14_+_56127989 | 0.50 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr6_-_109804412 | 0.50 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr8_+_104426942 | 0.49 |
ENST00000297579.5
|
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr14_+_56127960 | 0.49 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr6_+_27861190 | 0.49 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr6_+_109416684 | 0.49 |
ENST00000521522.1
ENST00000524064.1 ENST00000522608.1 ENST00000521503.1 ENST00000519407.1 ENST00000519095.1 ENST00000368968.2 ENST00000522490.1 ENST00000523209.1 ENST00000368970.2 ENST00000520883.1 ENST00000523787.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chrX_-_20159934 | 0.49 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr1_+_231473743 | 0.49 |
ENST00000295050.7
|
SPRTN
|
SprT-like N-terminal domain |
| chr6_+_57182400 | 0.48 |
ENST00000607273.1
|
PRIM2
|
primase, DNA, polypeptide 2 (58kDa) |
| chr10_-_38146510 | 0.48 |
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248 |
| chr5_-_40755987 | 0.48 |
ENST00000337702.4
|
TTC33
|
tetratricopeptide repeat domain 33 |
| chr5_+_33441053 | 0.48 |
ENST00000541634.1
ENST00000455217.2 ENST00000414361.2 |
TARS
|
threonyl-tRNA synthetase |
| chr21_+_40759684 | 0.48 |
ENST00000380708.1
|
WRB
|
tryptophan rich basic protein |
| chrX_-_11129229 | 0.48 |
ENST00000608176.1
ENST00000433747.2 ENST00000608576.1 ENST00000608916.1 |
RP11-120D5.1
|
RP11-120D5.1 |
| chr10_-_27389392 | 0.47 |
ENST00000376087.4
|
ANKRD26
|
ankyrin repeat domain 26 |
| chr13_+_38923959 | 0.47 |
ENST00000379649.1
ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1
|
ubiquitin-fold modifier 1 |
| chr3_+_142720366 | 0.47 |
ENST00000493782.1
ENST00000397933.2 ENST00000473835.2 ENST00000493598.2 |
U2SURP
|
U2 snRNP-associated SURP domain containing |
| chr5_+_93954039 | 0.47 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr2_-_231084617 | 0.47 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr1_-_101491319 | 0.47 |
ENST00000342173.7
ENST00000488176.1 ENST00000370109.3 |
DPH5
|
diphthamide biosynthesis 5 |
| chr1_+_207925391 | 0.46 |
ENST00000358170.2
ENST00000354848.1 ENST00000322918.5 ENST00000367042.1 ENST00000367041.1 ENST00000357714.1 ENST00000322875.4 ENST00000367047.1 ENST00000441839.2 ENST00000361067.1 ENST00000360212.2 ENST00000480003.1 |
CD46
|
CD46 molecule, complement regulatory protein |
| chr18_+_3247779 | 0.46 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr10_+_18948311 | 0.46 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr15_+_45879534 | 0.46 |
ENST00000564080.1
ENST00000562384.1 ENST00000569076.1 ENST00000566753.1 |
RP11-96O20.4
BLOC1S6
|
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr4_+_40058411 | 0.46 |
ENST00000261435.6
ENST00000515550.1 |
N4BP2
|
NEDD4 binding protein 2 |
| chr14_-_102552659 | 0.46 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr1_-_167883327 | 0.46 |
ENST00000476818.2
ENST00000367851.4 ENST00000367848.1 |
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr9_-_135230336 | 0.46 |
ENST00000224140.5
ENST00000372169.2 ENST00000393220.1 |
SETX
|
senataxin |
| chr19_+_58281014 | 0.46 |
ENST00000391702.3
ENST00000598885.1 ENST00000598183.1 ENST00000396154.2 ENST00000599802.1 ENST00000396150.4 |
ZNF586
|
zinc finger protein 586 |
| chr5_+_118604385 | 0.46 |
ENST00000274456.6
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr2_-_55920952 | 0.46 |
ENST00000447944.2
|
PNPT1
|
polyribonucleotide nucleotidyltransferase 1 |
| chr17_+_65713925 | 0.45 |
ENST00000253247.4
|
NOL11
|
nucleolar protein 11 |
| chr20_-_49575081 | 0.45 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr1_-_109203997 | 0.45 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr4_-_129209944 | 0.45 |
ENST00000520121.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr4_+_128802016 | 0.45 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr5_+_54455946 | 0.45 |
ENST00000503787.1
ENST00000296734.6 ENST00000515370.1 |
GPX8
|
glutathione peroxidase 8 (putative) |
| chr17_+_11924129 | 0.44 |
ENST00000353533.5
ENST00000415385.3 |
MAP2K4
|
mitogen-activated protein kinase kinase 4 |
| chr10_-_18940501 | 0.44 |
ENST00000377304.4
|
NSUN6
|
NOP2/Sun domain family, member 6 |
| chr9_+_33264861 | 0.44 |
ENST00000223500.8
|
CHMP5
|
charged multivesicular body protein 5 |
| chr14_+_31028348 | 0.44 |
ENST00000550944.1
ENST00000438909.2 ENST00000553504.1 |
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr6_+_56911476 | 0.43 |
ENST00000545356.1
|
KIAA1586
|
KIAA1586 |
| chr1_-_151342180 | 0.43 |
ENST00000424475.1
|
SELENBP1
|
selenium binding protein 1 |
| chr7_-_108168580 | 0.43 |
ENST00000453085.1
|
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr9_-_88896977 | 0.43 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr19_+_37960240 | 0.42 |
ENST00000388801.3
|
ZNF570
|
zinc finger protein 570 |
| chr1_-_236767779 | 0.42 |
ENST00000366579.1
ENST00000366582.3 ENST00000366581.2 |
HEATR1
|
HEAT repeat containing 1 |
| chr3_+_186501336 | 0.42 |
ENST00000323963.5
ENST00000440191.2 ENST00000356531.5 |
EIF4A2
|
eukaryotic translation initiation factor 4A2 |
| chr19_+_44331444 | 0.42 |
ENST00000324461.7
|
ZNF283
|
zinc finger protein 283 |
| chr6_+_109416313 | 0.42 |
ENST00000521277.1
ENST00000517392.1 ENST00000407272.1 ENST00000336977.4 ENST00000519286.1 ENST00000518329.1 ENST00000522461.1 ENST00000518853.1 |
CEP57L1
|
centrosomal protein 57kDa-like 1 |
| chr14_+_38033252 | 0.42 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr2_-_20251744 | 0.41 |
ENST00000175091.4
|
LAPTM4A
|
lysosomal protein transmembrane 4 alpha |
| chr15_-_55881135 | 0.41 |
ENST00000302000.6
|
PYGO1
|
pygopus family PHD finger 1 |
| chr10_-_18948156 | 0.41 |
ENST00000414939.1
ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr13_-_31736478 | 0.41 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr8_+_110552831 | 0.41 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr22_+_27068766 | 0.41 |
ENST00000435162.1
ENST00000437071.1 ENST00000440816.1 ENST00000421253.1 |
CTA-211A9.5
|
CTA-211A9.5 |
| chr12_-_64784306 | 0.41 |
ENST00000543259.1
|
C12orf56
|
chromosome 12 open reading frame 56 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SP100
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.6 | 1.9 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.5 | 1.5 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.3 | 0.3 | GO:1902956 | negative regulation of cellular respiration(GO:1901856) regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.3 | 0.8 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.2 | 1.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.2 | 0.7 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.2 | 0.9 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 0.6 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.2 | 1.6 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.2 | 1.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 1.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.2 | 1.4 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.2 | 0.5 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.2 | 0.5 | GO:2000627 | rRNA import into mitochondrion(GO:0035928) regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.1 | 0.7 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 1.2 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 1.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 1.0 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.5 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.6 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.7 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.4 | GO:0061508 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 0.6 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.8 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.4 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 0.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.4 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.0 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.3 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.1 | 0.3 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.4 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.5 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.4 | GO:2000672 | cellular response to sorbitol(GO:0072709) negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 1.4 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.4 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.6 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:0015883 | FAD transport(GO:0015883) FAD transmembrane transport(GO:0035350) |
| 0.1 | 1.0 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 2.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.1 | 0.8 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.1 | 0.4 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.1 | 1.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 1.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.8 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.8 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 0.3 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.1 | 0.2 | GO:2000987 | cell communication by chemical coupling(GO:0010643) positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.3 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.2 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 1.1 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 0.4 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.1 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 1.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.3 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.1 | 0.4 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.2 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.1 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 1.8 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.1 | 0.5 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.1 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.9 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.0 | 0.2 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 1.6 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.9 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.6 | GO:0071316 | cellular response to nicotine(GO:0071316) |
| 0.0 | 0.8 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.7 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.6 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.5 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 1.0 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.6 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.9 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.4 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.6 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.4 | GO:2001171 | positive regulation of ATP biosynthetic process(GO:2001171) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 1.8 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 1.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.3 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:2000691 | regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.3 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.6 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.3 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 | 0.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 1.2 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:1904871 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.2 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.0 | 0.2 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.5 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.6 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 1.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.6 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.4 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.2 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.6 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.6 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.0 | 0.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.0 | 0.3 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.6 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.3 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.1 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.5 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.5 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.7 | GO:0007077 | mitotic nuclear envelope disassembly(GO:0007077) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.4 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.4 | 1.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.3 | 0.9 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.2 | 0.6 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.2 | 0.7 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 0.8 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 0.8 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.9 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 3.1 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.6 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.7 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 1.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.7 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.5 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.9 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 0.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 1.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.4 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 3.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.3 | 0.9 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 1.5 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 1.0 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 0.9 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 0.6 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.2 | 0.6 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.2 | 0.6 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.2 | 0.8 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.2 | 0.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 1.3 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.2 | 0.7 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 0.8 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 0.6 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.2 | 0.8 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.2 | 0.6 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.2 | 0.5 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.6 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 1.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.6 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.1 | 0.5 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.8 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.6 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.1 | 0.5 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 0.8 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.3 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 0.4 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 1.4 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.2 | GO:0051908 | double-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0051908) |
| 0.1 | 0.3 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 0.6 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 1.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.6 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.1 | 0.4 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.1 | 0.4 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 2.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.5 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.2 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 1.0 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.4 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.3 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.4 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.4 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0016768 | spermine synthase activity(GO:0016768) |
| 0.0 | 0.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.4 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 1.0 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 2.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.4 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.2 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.4 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 1.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 2.5 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.3 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.2 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.0 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 2.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.5 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 1.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.1 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.1 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.1 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 4.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.6 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |