Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
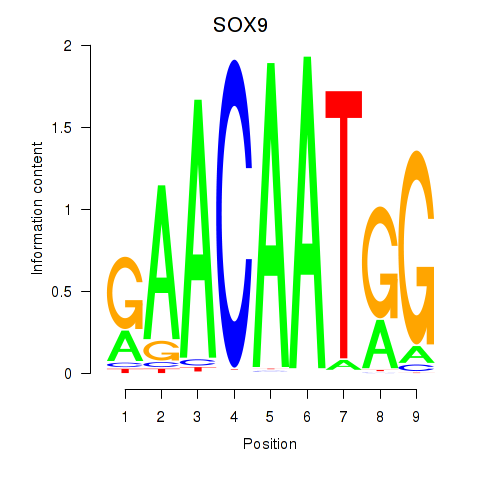
Results for SOX9
Z-value: 0.56
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.5 | SRY-box transcription factor 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX9 | hg19_v2_chr17_+_70117153_70117174 | 0.33 | 5.2e-01 | Click! |
Activity profile of SOX9 motif
Sorted Z-values of SOX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_175204865 | 0.36 |
ENST00000505124.1
|
CEP44
|
centrosomal protein 44kDa |
| chr16_+_82660635 | 0.33 |
ENST00000567445.1
ENST00000446376.2 |
CDH13
|
cadherin 13 |
| chr3_-_122283424 | 0.32 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr11_+_28724129 | 0.28 |
ENST00000513853.1
|
RP11-115J23.1
|
RP11-115J23.1 |
| chr14_+_24641062 | 0.27 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr1_+_26437631 | 0.27 |
ENST00000444713.1
|
PDIK1L
|
PDLIM1 interacting kinase 1 like |
| chr7_+_77167343 | 0.26 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr7_+_77167376 | 0.24 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr17_-_14683517 | 0.23 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr14_-_51027838 | 0.23 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr1_-_156399184 | 0.21 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr1_-_27339317 | 0.21 |
ENST00000289166.5
|
FAM46B
|
family with sequence similarity 46, member B |
| chr8_+_126442563 | 0.21 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr1_+_181003067 | 0.20 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr7_-_99774945 | 0.19 |
ENST00000292377.2
|
GPC2
|
glypican 2 |
| chr11_+_34654011 | 0.19 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr3_+_189507460 | 0.19 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr6_+_26183958 | 0.19 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr19_-_17375541 | 0.19 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr11_+_110225855 | 0.19 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr7_-_107968999 | 0.19 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr18_+_44497455 | 0.17 |
ENST00000592005.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr14_-_71276211 | 0.16 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr7_-_107968921 | 0.16 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr12_-_49463620 | 0.16 |
ENST00000550675.1
|
RHEBL1
|
Ras homolog enriched in brain like 1 |
| chr6_+_29691198 | 0.16 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr8_-_121824374 | 0.16 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr6_+_29691056 | 0.15 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr9_-_85882145 | 0.15 |
ENST00000328788.1
|
FRMD3
|
FERM domain containing 3 |
| chr7_+_129015671 | 0.15 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr2_-_37501692 | 0.14 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr1_-_16539094 | 0.14 |
ENST00000270747.3
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr5_-_168006324 | 0.14 |
ENST00000522176.1
|
PANK3
|
pantothenate kinase 3 |
| chr4_+_71570430 | 0.14 |
ENST00000417478.2
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr18_+_7946839 | 0.14 |
ENST00000578916.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr11_-_77531752 | 0.14 |
ENST00000440064.2
ENST00000528095.1 |
RSF1
|
remodeling and spacing factor 1 |
| chr10_+_121578211 | 0.13 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr11_-_65640071 | 0.13 |
ENST00000526624.1
|
EFEMP2
|
EGF containing fibulin-like extracellular matrix protein 2 |
| chr8_-_66753682 | 0.13 |
ENST00000396642.3
|
PDE7A
|
phosphodiesterase 7A |
| chr9_-_74383302 | 0.13 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr2_+_208576259 | 0.13 |
ENST00000392209.3
|
CCNYL1
|
cyclin Y-like 1 |
| chr14_-_90421028 | 0.13 |
ENST00000267544.9
ENST00000316738.7 ENST00000538485.2 ENST00000556609.1 |
EFCAB11
|
EF-hand calcium binding domain 11 |
| chr16_-_15474904 | 0.12 |
ENST00000534094.1
|
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr3_-_185826718 | 0.12 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr15_+_41136216 | 0.12 |
ENST00000562057.1
ENST00000344051.4 |
SPINT1
|
serine peptidase inhibitor, Kunitz type 1 |
| chr20_+_42543504 | 0.12 |
ENST00000341197.4
|
TOX2
|
TOX high mobility group box family member 2 |
| chr12_+_49761147 | 0.12 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr6_-_27440460 | 0.12 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr12_+_95611569 | 0.12 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr19_-_19739321 | 0.12 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr4_+_54243917 | 0.12 |
ENST00000507166.1
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr8_-_95449155 | 0.11 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr4_+_54243862 | 0.11 |
ENST00000306932.6
|
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr20_+_60698180 | 0.11 |
ENST00000361670.3
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae) |
| chr9_+_4792869 | 0.11 |
ENST00000381750.4
|
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr2_+_46926326 | 0.11 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr6_-_27440837 | 0.11 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr12_-_99548524 | 0.11 |
ENST00000549558.2
ENST00000550693.2 ENST00000549493.2 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_-_139658965 | 0.11 |
ENST00000316144.5
|
LCN15
|
lipocalin 15 |
| chr12_+_69633407 | 0.11 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr17_-_73149921 | 0.11 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chrX_+_9433289 | 0.11 |
ENST00000422314.1
|
TBL1X
|
transducin (beta)-like 1X-linked |
| chr22_-_38480100 | 0.11 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr2_+_46926048 | 0.11 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr12_+_95611516 | 0.11 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr15_-_59041954 | 0.10 |
ENST00000439637.1
ENST00000558004.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr6_-_75915757 | 0.10 |
ENST00000322507.8
|
COL12A1
|
collagen, type XII, alpha 1 |
| chr18_+_7946941 | 0.10 |
ENST00000444013.1
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M |
| chr7_+_76751926 | 0.10 |
ENST00000285871.4
ENST00000431197.1 |
CCDC146
|
coiled-coil domain containing 146 |
| chr5_-_65018834 | 0.10 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr15_+_98503922 | 0.10 |
ENST00000268042.6
|
ARRDC4
|
arrestin domain containing 4 |
| chr4_+_113152978 | 0.10 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr2_+_54198210 | 0.10 |
ENST00000607452.1
ENST00000422521.2 |
ACYP2
|
acylphosphatase 2, muscle type |
| chr17_-_16472483 | 0.10 |
ENST00000395824.1
ENST00000448349.2 ENST00000395825.3 |
ZNF287
|
zinc finger protein 287 |
| chr8_+_98656693 | 0.10 |
ENST00000519934.1
|
MTDH
|
metadherin |
| chr2_-_175869936 | 0.10 |
ENST00000409900.3
|
CHN1
|
chimerin 1 |
| chr6_-_134861089 | 0.10 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr13_-_41593425 | 0.10 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr7_+_134464376 | 0.10 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr1_-_243418650 | 0.10 |
ENST00000522995.1
|
CEP170
|
centrosomal protein 170kDa |
| chr20_-_17662878 | 0.10 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chr5_+_138852090 | 0.10 |
ENST00000593907.1
|
AC138517.1
|
Uncharacterized protein |
| chr16_+_68573116 | 0.10 |
ENST00000570495.1
ENST00000563169.2 ENST00000564323.1 ENST00000562156.1 ENST00000573685.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr2_-_201828356 | 0.10 |
ENST00000234296.2
|
ORC2
|
origin recognition complex, subunit 2 |
| chr4_-_83351294 | 0.10 |
ENST00000502762.1
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr11_+_111807863 | 0.09 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr1_+_8378140 | 0.09 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr19_+_58790314 | 0.09 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr13_-_107214291 | 0.09 |
ENST00000375926.1
|
ARGLU1
|
arginine and glutamate rich 1 |
| chr12_+_49761224 | 0.09 |
ENST00000553127.1
ENST00000321898.6 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr1_-_198906528 | 0.09 |
ENST00000432296.1
|
MIR181A1HG
|
MIR181A1 host gene (non-protein coding) |
| chr5_+_110073853 | 0.09 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr11_-_76155618 | 0.09 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr17_+_40119801 | 0.09 |
ENST00000585452.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr17_+_9066252 | 0.09 |
ENST00000436734.1
|
NTN1
|
netrin 1 |
| chr1_-_184723701 | 0.09 |
ENST00000367512.3
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr10_-_104953009 | 0.09 |
ENST00000470299.1
ENST00000343289.5 |
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr4_+_41614720 | 0.09 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_-_205912577 | 0.09 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chrX_+_135579670 | 0.09 |
ENST00000218364.4
|
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr2_-_70475586 | 0.09 |
ENST00000416149.2
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr7_-_75368248 | 0.09 |
ENST00000434438.2
ENST00000336926.6 |
HIP1
|
huntingtin interacting protein 1 |
| chr3_+_44596679 | 0.09 |
ENST00000426540.1
ENST00000431636.1 ENST00000341840.3 ENST00000273320.3 |
ZKSCAN7
|
zinc finger with KRAB and SCAN domains 7 |
| chr15_+_90895471 | 0.09 |
ENST00000354377.3
ENST00000379090.5 |
ZNF774
|
zinc finger protein 774 |
| chr4_-_83350580 | 0.09 |
ENST00000349655.4
ENST00000602300.1 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr17_+_57233087 | 0.09 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr4_+_54243798 | 0.09 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chrX_+_9880412 | 0.09 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr1_+_209602156 | 0.09 |
ENST00000429156.1
ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr10_+_70748487 | 0.09 |
ENST00000361983.4
|
KIAA1279
|
KIAA1279 |
| chr15_+_76629064 | 0.08 |
ENST00000290759.4
|
ISL2
|
ISL LIM homeobox 2 |
| chr12_+_113495492 | 0.08 |
ENST00000257600.3
|
DTX1
|
deltex homolog 1 (Drosophila) |
| chr5_+_138678131 | 0.08 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr1_-_182360498 | 0.08 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr7_-_27219632 | 0.08 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr19_+_44100632 | 0.08 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr1_-_205904950 | 0.08 |
ENST00000340781.4
|
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr2_+_170590321 | 0.08 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr19_-_45004556 | 0.08 |
ENST00000587047.1
ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180
|
zinc finger protein 180 |
| chr12_-_95945246 | 0.08 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chr7_-_16685422 | 0.08 |
ENST00000306999.2
|
ANKMY2
|
ankyrin repeat and MYND domain containing 2 |
| chr11_+_13299186 | 0.08 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr6_+_22146851 | 0.08 |
ENST00000606197.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr15_-_59041768 | 0.08 |
ENST00000402627.1
ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10
|
ADAM metallopeptidase domain 10 |
| chr14_-_91884150 | 0.08 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr20_-_1317555 | 0.08 |
ENST00000537552.1
|
AL136531.1
|
HCG2043693; Uncharacterized protein |
| chr1_-_40367668 | 0.08 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr5_+_140767452 | 0.08 |
ENST00000519479.1
|
PCDHGB4
|
protocadherin gamma subfamily B, 4 |
| chr4_+_175205162 | 0.08 |
ENST00000503053.1
|
CEP44
|
centrosomal protein 44kDa |
| chr19_+_16187816 | 0.07 |
ENST00000588410.1
|
TPM4
|
tropomyosin 4 |
| chr12_-_95611149 | 0.07 |
ENST00000549499.1
ENST00000343958.4 ENST00000546711.1 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr15_+_89922345 | 0.07 |
ENST00000558982.1
|
LINC00925
|
long intergenic non-protein coding RNA 925 |
| chr10_-_62332357 | 0.07 |
ENST00000503366.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr8_+_133787586 | 0.07 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr12_+_69979210 | 0.07 |
ENST00000544368.2
|
CCT2
|
chaperonin containing TCP1, subunit 2 (beta) |
| chr1_+_16085244 | 0.07 |
ENST00000400773.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr20_-_17662705 | 0.07 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr11_+_63953691 | 0.07 |
ENST00000543847.1
|
STIP1
|
stress-induced-phosphoprotein 1 |
| chr17_-_42441204 | 0.07 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr6_-_79944336 | 0.07 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr8_+_42196000 | 0.07 |
ENST00000518925.1
ENST00000538005.1 |
POLB
|
polymerase (DNA directed), beta |
| chr19_+_18208603 | 0.07 |
ENST00000262811.6
|
MAST3
|
microtubule associated serine/threonine kinase 3 |
| chr1_-_243418344 | 0.07 |
ENST00000366542.1
|
CEP170
|
centrosomal protein 170kDa |
| chr15_-_52263937 | 0.07 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr9_+_103235365 | 0.07 |
ENST00000374879.4
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr1_-_153643442 | 0.07 |
ENST00000368681.1
ENST00000361891.4 |
ILF2
|
interleukin enhancer binding factor 2 |
| chr3_-_128294929 | 0.07 |
ENST00000356020.2
|
C3orf27
|
chromosome 3 open reading frame 27 |
| chr1_+_185014496 | 0.07 |
ENST00000367510.3
|
RNF2
|
ring finger protein 2 |
| chr15_+_57211318 | 0.07 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr11_-_102323489 | 0.07 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr14_-_38036271 | 0.07 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr12_-_120805872 | 0.07 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr6_+_41040678 | 0.07 |
ENST00000341376.6
ENST00000353205.5 |
NFYA
|
nuclear transcription factor Y, alpha |
| chr14_+_75230011 | 0.07 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chr2_-_37899323 | 0.07 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr17_+_67498295 | 0.07 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr15_+_71184931 | 0.07 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr12_+_49761273 | 0.07 |
ENST00000551540.1
ENST00000552918.1 ENST00000548777.1 ENST00000547865.1 ENST00000552171.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr5_-_131826457 | 0.07 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr1_+_33116765 | 0.07 |
ENST00000544435.1
ENST00000373485.1 ENST00000458695.2 ENST00000490500.1 ENST00000445722.2 |
RBBP4
|
retinoblastoma binding protein 4 |
| chr7_+_12726623 | 0.06 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr18_-_24128496 | 0.06 |
ENST00000417602.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr12_-_118797475 | 0.06 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr6_+_32132360 | 0.06 |
ENST00000333845.6
ENST00000395512.1 ENST00000432129.1 |
EGFL8
|
EGF-like-domain, multiple 8 |
| chr18_+_20494078 | 0.06 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr12_-_12674032 | 0.06 |
ENST00000298573.4
|
DUSP16
|
dual specificity phosphatase 16 |
| chr20_-_34330129 | 0.06 |
ENST00000397370.3
ENST00000528062.3 ENST00000407261.4 ENST00000374038.3 ENST00000361162.6 |
RBM39
|
RNA binding motif protein 39 |
| chr4_-_83351005 | 0.06 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr1_-_200992827 | 0.06 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr7_+_16793160 | 0.06 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr17_+_7748233 | 0.06 |
ENST00000570632.1
|
KDM6B
|
lysine (K)-specific demethylase 6B |
| chr4_-_5894777 | 0.06 |
ENST00000324989.7
|
CRMP1
|
collapsin response mediator protein 1 |
| chr16_+_14802801 | 0.06 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr12_+_49212514 | 0.06 |
ENST00000301050.2
ENST00000548279.1 ENST00000547230.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr14_-_69619291 | 0.06 |
ENST00000554215.1
ENST00000556847.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr12_-_2966193 | 0.06 |
ENST00000382678.3
|
AC005841.1
|
Uncharacterized protein ENSP00000372125 |
| chr7_+_26191809 | 0.06 |
ENST00000056233.3
|
NFE2L3
|
nuclear factor, erythroid 2-like 3 |
| chr12_+_95611536 | 0.06 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr17_-_56605341 | 0.06 |
ENST00000583114.1
|
SEPT4
|
septin 4 |
| chr20_-_45984401 | 0.06 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr1_-_184723942 | 0.06 |
ENST00000318130.8
|
EDEM3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr9_+_4792971 | 0.06 |
ENST00000381732.3
ENST00000442869.1 |
RCL1
|
RNA terminal phosphate cyclase-like 1 |
| chr7_+_11013491 | 0.06 |
ENST00000403050.3
ENST00000445996.2 |
PHF14
|
PHD finger protein 14 |
| chr14_-_88459182 | 0.06 |
ENST00000544807.2
|
GALC
|
galactosylceramidase |
| chr2_+_102314161 | 0.06 |
ENST00000425019.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr11_-_111649015 | 0.06 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chrX_+_49126294 | 0.06 |
ENST00000466508.1
ENST00000438316.1 ENST00000055335.6 ENST00000495799.1 |
PPP1R3F
|
protein phosphatase 1, regulatory subunit 3F |
| chr8_+_42195972 | 0.06 |
ENST00000532157.1
ENST00000265421.4 ENST00000520008.1 |
POLB
|
polymerase (DNA directed), beta |
| chr5_-_179780312 | 0.06 |
ENST00000253778.8
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr14_-_88459503 | 0.06 |
ENST00000393568.4
ENST00000261304.2 |
GALC
|
galactosylceramidase |
| chr6_-_15586238 | 0.06 |
ENST00000462989.2
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chrX_-_40005865 | 0.06 |
ENST00000412952.1
|
BCOR
|
BCL6 corepressor |
| chr14_+_67291158 | 0.06 |
ENST00000555456.1
|
GPHN
|
gephyrin |
| chr6_+_27100811 | 0.06 |
ENST00000359193.2
|
HIST1H2AG
|
histone cluster 1, H2ag |
| chr11_+_125496619 | 0.06 |
ENST00000532669.1
ENST00000278916.3 |
CHEK1
|
checkpoint kinase 1 |
| chr1_-_26232951 | 0.06 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr22_-_30970560 | 0.06 |
ENST00000402369.1
ENST00000406361.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr15_+_91416092 | 0.06 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chrX_+_107068959 | 0.06 |
ENST00000451923.1
|
MID2
|
midline 2 |
| chr10_-_120101804 | 0.06 |
ENST00000369183.4
ENST00000369172.4 |
FAM204A
|
family with sequence similarity 204, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.0 | GO:0035787 | cell migration involved in kidney development(GO:0035787) |
| 0.0 | 0.3 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.2 | GO:0045658 | eosinophil differentiation(GO:0030222) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.2 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.0 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.5 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.0 | 0.1 | GO:0035740 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:2000584 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.0 | GO:1902275 | regulation of chromosome organization(GO:0033044) regulation of chromatin organization(GO:1902275) regulation of chromatin modification(GO:1903308) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0072579 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.1 | GO:0032900 | negative regulation of neurotrophin production(GO:0032900) |
| 0.0 | 0.2 | GO:0035977 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.1 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.3 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.0 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.3 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.1 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.0 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.0 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |