Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
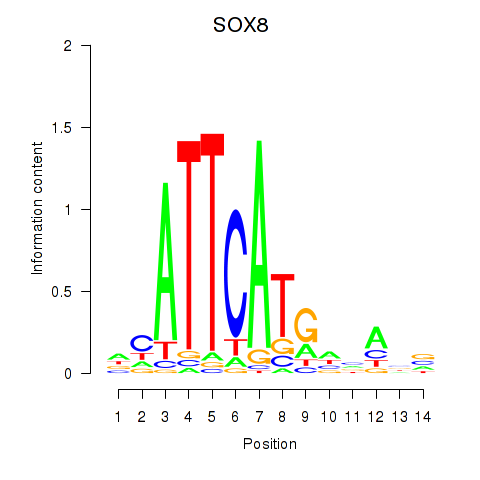
Results for SOX8
Z-value: 0.46
Transcription factors associated with SOX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX8
|
ENSG00000005513.9 | SRY-box transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX8 | hg19_v2_chr16_+_1031762_1031808 | 0.28 | 5.9e-01 | Click! |
Activity profile of SOX8 motif
Sorted Z-values of SOX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_58204128 | 0.29 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr2_+_7118755 | 0.27 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr19_-_21950332 | 0.27 |
ENST00000598026.1
|
ZNF100
|
zinc finger protein 100 |
| chr1_+_81106951 | 0.26 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr2_-_36779411 | 0.24 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr12_-_31158902 | 0.23 |
ENST00000544329.1
ENST00000418254.2 ENST00000222396.5 |
RP11-551L14.4
|
RP11-551L14.4 |
| chr14_-_92247032 | 0.22 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr3_-_187455680 | 0.21 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr12_-_10282836 | 0.21 |
ENST00000304084.8
ENST00000353231.5 ENST00000525605.1 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr17_+_68047418 | 0.20 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr12_+_78359999 | 0.18 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr5_+_156887027 | 0.18 |
ENST00000435489.2
ENST00000311946.7 |
NIPAL4
|
NIPA-like domain containing 4 |
| chr9_-_128246769 | 0.18 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr12_-_120241187 | 0.18 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr10_-_375422 | 0.17 |
ENST00000434695.2
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr7_+_100303676 | 0.16 |
ENST00000303151.4
|
POP7
|
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr6_+_42018614 | 0.16 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr2_-_14541060 | 0.15 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr12_-_10282742 | 0.15 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr3_-_165555200 | 0.15 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr3_+_11267691 | 0.15 |
ENST00000413416.1
|
HRH1
|
histamine receptor H1 |
| chr16_-_28503357 | 0.15 |
ENST00000333496.9
ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr22_+_45714301 | 0.15 |
ENST00000427777.1
|
FAM118A
|
family with sequence similarity 118, member A |
| chr9_-_139891165 | 0.14 |
ENST00000494426.1
|
CLIC3
|
chloride intracellular channel 3 |
| chr16_+_30076052 | 0.14 |
ENST00000563987.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr3_-_100558953 | 0.14 |
ENST00000533795.1
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr20_+_42543504 | 0.13 |
ENST00000341197.4
|
TOX2
|
TOX high mobility group box family member 2 |
| chr9_-_95244781 | 0.13 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr4_+_22999152 | 0.12 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr5_-_146781153 | 0.12 |
ENST00000520473.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr15_-_65715401 | 0.12 |
ENST00000352385.2
|
IGDCC4
|
immunoglobulin superfamily, DCC subclass, member 4 |
| chr20_-_1472029 | 0.12 |
ENST00000359801.3
|
SIRPB2
|
signal-regulatory protein beta 2 |
| chr17_+_36584662 | 0.12 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr5_+_175288631 | 0.12 |
ENST00000509837.1
|
CPLX2
|
complexin 2 |
| chr4_-_137020391 | 0.11 |
ENST00000569694.1
|
RP11-775H9.3
|
RP11-775H9.3 |
| chr17_+_7387677 | 0.11 |
ENST00000322644.6
|
POLR2A
|
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr11_-_9482010 | 0.11 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr16_+_87636474 | 0.11 |
ENST00000284262.2
|
JPH3
|
junctophilin 3 |
| chr1_+_110577229 | 0.11 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr6_-_136788001 | 0.11 |
ENST00000544465.1
|
MAP7
|
microtubule-associated protein 7 |
| chr1_+_20959943 | 0.11 |
ENST00000321556.4
|
PINK1
|
PTEN induced putative kinase 1 |
| chr19_+_49535169 | 0.11 |
ENST00000474913.1
ENST00000359342.6 |
CGB2
|
chorionic gonadotropin, beta polypeptide 2 |
| chr1_-_183538319 | 0.11 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr2_-_70944855 | 0.10 |
ENST00000415348.1
|
ADD2
|
adducin 2 (beta) |
| chr20_-_5426380 | 0.10 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr17_-_47723943 | 0.10 |
ENST00000510476.1
ENST00000503676.1 |
SPOP
|
speckle-type POZ protein |
| chr15_-_72410455 | 0.10 |
ENST00000569314.1
|
MYO9A
|
myosin IXA |
| chr6_-_27100529 | 0.10 |
ENST00000607124.1
ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ
|
histone cluster 1, H2bj |
| chr19_+_19144384 | 0.10 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr1_-_145715565 | 0.10 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr2_+_102615416 | 0.10 |
ENST00000393414.2
|
IL1R2
|
interleukin 1 receptor, type II |
| chr18_-_25616519 | 0.10 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr18_+_11857439 | 0.10 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr17_+_41132564 | 0.10 |
ENST00000361677.1
ENST00000589705.1 |
RUNDC1
|
RUN domain containing 1 |
| chr10_-_74114714 | 0.10 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr18_+_61575200 | 0.10 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr4_-_47983519 | 0.09 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr12_-_49581152 | 0.09 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr6_+_37012607 | 0.09 |
ENST00000423336.1
|
COX6A1P2
|
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
| chr12_+_79371565 | 0.09 |
ENST00000551304.1
|
SYT1
|
synaptotagmin I |
| chrX_+_48687283 | 0.09 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr12_+_21207503 | 0.09 |
ENST00000545916.1
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr10_-_101825151 | 0.09 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr16_+_30075783 | 0.09 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr2_+_13677795 | 0.09 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr16_+_21244986 | 0.09 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr17_-_7145475 | 0.09 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr17_-_27230035 | 0.09 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr1_+_46269248 | 0.09 |
ENST00000361297.2
ENST00000372009.2 |
MAST2
|
microtubule associated serine/threonine kinase 2 |
| chr2_+_233527443 | 0.09 |
ENST00000410095.1
|
EFHD1
|
EF-hand domain family, member D1 |
| chr19_-_47287990 | 0.09 |
ENST00000593713.1
ENST00000598022.1 ENST00000434726.2 |
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr2_-_37501692 | 0.08 |
ENST00000443977.1
|
PRKD3
|
protein kinase D3 |
| chr2_+_39117010 | 0.08 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr16_+_57769635 | 0.08 |
ENST00000379661.3
ENST00000562592.1 ENST00000566726.1 |
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr6_+_31707725 | 0.08 |
ENST00000375755.3
ENST00000375742.3 ENST00000375750.3 ENST00000425703.1 ENST00000534153.4 ENST00000375703.3 ENST00000375740.3 |
MSH5
|
mutS homolog 5 |
| chr11_-_33743952 | 0.08 |
ENST00000534312.1
|
CD59
|
CD59 molecule, complement regulatory protein |
| chr22_-_36903069 | 0.08 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr10_-_6104253 | 0.08 |
ENST00000256876.6
ENST00000379954.1 ENST00000379959.3 |
IL2RA
|
interleukin 2 receptor, alpha |
| chr8_+_7752151 | 0.08 |
ENST00000302247.2
|
DEFB4A
|
defensin, beta 4A |
| chr12_-_123187890 | 0.08 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr7_-_64467031 | 0.08 |
ENST00000394323.2
|
ERV3-1
|
endogenous retrovirus group 3, member 1 |
| chr12_+_133066137 | 0.08 |
ENST00000434748.2
|
FBRSL1
|
fibrosin-like 1 |
| chr1_-_152297679 | 0.08 |
ENST00000368799.1
|
FLG
|
filaggrin |
| chr12_+_122242597 | 0.08 |
ENST00000267197.5
|
SETD1B
|
SET domain containing 1B |
| chr3_-_123339418 | 0.08 |
ENST00000583087.1
|
MYLK
|
myosin light chain kinase |
| chr15_+_80733570 | 0.08 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr17_-_73663245 | 0.08 |
ENST00000584999.1
ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5
|
RecQ protein-like 5 |
| chr16_+_30075595 | 0.07 |
ENST00000563060.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr16_+_30075463 | 0.07 |
ENST00000562168.1
ENST00000569545.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr15_+_91445448 | 0.07 |
ENST00000558290.1
ENST00000558853.1 ENST00000559999.1 |
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr12_+_48876275 | 0.07 |
ENST00000314014.2
|
C12orf54
|
chromosome 12 open reading frame 54 |
| chr14_-_101053739 | 0.07 |
ENST00000554140.1
|
BEGAIN
|
brain-enriched guanylate kinase-associated |
| chr5_+_140602904 | 0.07 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr22_-_30722912 | 0.07 |
ENST00000215790.7
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr4_+_156680153 | 0.07 |
ENST00000502959.1
ENST00000505764.1 ENST00000507146.1 ENST00000264424.8 ENST00000503520.1 |
GUCY1B3
|
guanylate cyclase 1, soluble, beta 3 |
| chr21_-_43735628 | 0.07 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr3_-_47934234 | 0.07 |
ENST00000420772.2
|
MAP4
|
microtubule-associated protein 4 |
| chr16_+_14280742 | 0.07 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr10_-_116418053 | 0.07 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr5_+_140186647 | 0.07 |
ENST00000512229.2
ENST00000356878.4 ENST00000530339.1 |
PCDHA4
|
protocadherin alpha 4 |
| chr14_-_69263043 | 0.07 |
ENST00000408913.2
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_-_200992827 | 0.07 |
ENST00000332129.2
ENST00000422435.2 |
KIF21B
|
kinesin family member 21B |
| chr19_+_18794470 | 0.07 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr14_-_21562648 | 0.07 |
ENST00000555270.1
|
ZNF219
|
zinc finger protein 219 |
| chr22_-_30642728 | 0.07 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr22_-_30722866 | 0.06 |
ENST00000403477.3
|
TBC1D10A
|
TBC1 domain family, member 10A |
| chr1_+_26496362 | 0.06 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr17_-_66951382 | 0.06 |
ENST00000586539.1
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr12_-_123201337 | 0.06 |
ENST00000528880.2
|
HCAR3
|
hydroxycarboxylic acid receptor 3 |
| chr5_+_140739537 | 0.06 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr10_-_81932771 | 0.06 |
ENST00000437799.1
|
ANXA11
|
annexin A11 |
| chr8_+_21915368 | 0.06 |
ENST00000265800.5
ENST00000517418.1 |
DMTN
|
dematin actin binding protein |
| chr16_+_31271274 | 0.06 |
ENST00000287497.8
ENST00000544665.3 |
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr5_+_140753444 | 0.06 |
ENST00000517434.1
|
PCDHGA6
|
protocadherin gamma subfamily A, 6 |
| chr1_+_35734616 | 0.06 |
ENST00000441447.1
|
ZMYM4
|
zinc finger, MYM-type 4 |
| chr16_+_87985029 | 0.06 |
ENST00000439677.1
ENST00000286122.7 ENST00000355163.5 ENST00000454563.1 ENST00000479780.2 ENST00000393208.2 ENST00000412691.1 ENST00000355022.4 |
BANP
|
BTG3 associated nuclear protein |
| chr15_+_81299370 | 0.06 |
ENST00000560091.1
|
C15orf26
|
chromosome 15 open reading frame 26 |
| chr17_+_1646130 | 0.06 |
ENST00000453066.1
ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr2_+_7073174 | 0.06 |
ENST00000416587.1
|
RNF144A
|
ring finger protein 144A |
| chr6_-_138539627 | 0.06 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr3_-_52090461 | 0.06 |
ENST00000296483.6
ENST00000495880.1 |
DUSP7
|
dual specificity phosphatase 7 |
| chr19_+_51728316 | 0.06 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr17_-_41132410 | 0.06 |
ENST00000409446.3
ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr20_+_32150140 | 0.06 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr7_-_47578840 | 0.06 |
ENST00000450444.1
|
TNS3
|
tensin 3 |
| chr2_-_190448118 | 0.06 |
ENST00000440626.1
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr14_-_69262789 | 0.06 |
ENST00000557022.1
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr17_-_66951474 | 0.06 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr9_+_127624387 | 0.06 |
ENST00000353214.2
|
ARPC5L
|
actin related protein 2/3 complex, subunit 5-like |
| chrX_+_41548220 | 0.06 |
ENST00000378142.4
|
GPR34
|
G protein-coupled receptor 34 |
| chr6_-_131211534 | 0.06 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr4_-_89442940 | 0.06 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr19_-_45681482 | 0.05 |
ENST00000592647.1
ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A
|
trafficking protein particle complex 6A |
| chr17_-_43339474 | 0.05 |
ENST00000331780.4
|
SPATA32
|
spermatogenesis associated 32 |
| chrX_-_53461305 | 0.05 |
ENST00000168216.6
|
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr12_+_14927270 | 0.05 |
ENST00000544848.1
|
H2AFJ
|
H2A histone family, member J |
| chrX_+_114874727 | 0.05 |
ENST00000543070.1
|
PLS3
|
plastin 3 |
| chr3_+_49711391 | 0.05 |
ENST00000296456.5
ENST00000449966.1 |
APEH
|
acylaminoacyl-peptide hydrolase |
| chr6_+_148593425 | 0.05 |
ENST00000367469.1
|
SASH1
|
SAM and SH3 domain containing 1 |
| chr17_-_7387524 | 0.05 |
ENST00000311403.4
|
ZBTB4
|
zinc finger and BTB domain containing 4 |
| chr5_-_89770582 | 0.05 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr15_-_72563585 | 0.05 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr2_+_166326157 | 0.05 |
ENST00000421875.1
ENST00000314499.7 ENST00000409664.1 |
CSRNP3
|
cysteine-serine-rich nuclear protein 3 |
| chr3_+_157154578 | 0.05 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chrX_+_15767971 | 0.05 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chrX_-_53461288 | 0.05 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr5_-_138534071 | 0.05 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr19_+_36606933 | 0.05 |
ENST00000586868.1
|
TBCB
|
tubulin folding cofactor B |
| chr11_-_62995986 | 0.05 |
ENST00000403374.2
|
SLC22A25
|
solute carrier family 22, member 25 |
| chr3_-_176914191 | 0.05 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr22_-_20255212 | 0.05 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr19_-_42806842 | 0.05 |
ENST00000596265.1
|
PAFAH1B3
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr4_+_95972822 | 0.05 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr9_+_97766409 | 0.05 |
ENST00000425634.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr2_+_158114051 | 0.05 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr13_+_98794810 | 0.05 |
ENST00000595437.1
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr22_+_24236191 | 0.05 |
ENST00000215754.7
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr5_-_94417314 | 0.05 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr14_+_52327350 | 0.05 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr2_+_74011316 | 0.05 |
ENST00000409561.1
|
C2orf78
|
chromosome 2 open reading frame 78 |
| chr17_+_2240916 | 0.05 |
ENST00000574563.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr2_+_219646462 | 0.05 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr20_-_44176060 | 0.05 |
ENST00000354280.4
ENST00000504988.1 |
EPPIN
EPPIN-WFDC6
|
epididymal peptidase inhibitor EPPIN-WFDC6 readthrough |
| chr5_-_78809950 | 0.04 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr4_+_169633310 | 0.04 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr3_-_129147432 | 0.04 |
ENST00000503957.1
ENST00000505956.1 ENST00000326085.3 |
EFCAB12
|
EF-hand calcium binding domain 12 |
| chr1_+_8378140 | 0.04 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr12_-_15103621 | 0.04 |
ENST00000536592.1
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr8_-_135522425 | 0.04 |
ENST00000521673.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr11_+_10477733 | 0.04 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr16_-_122619 | 0.04 |
ENST00000262316.6
|
RHBDF1
|
rhomboid 5 homolog 1 (Drosophila) |
| chr5_-_160973649 | 0.04 |
ENST00000393959.1
ENST00000517547.1 |
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr5_+_140792614 | 0.04 |
ENST00000398610.2
|
PCDHGA10
|
protocadherin gamma subfamily A, 10 |
| chr20_+_48807351 | 0.04 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr21_-_19858196 | 0.04 |
ENST00000422787.1
|
TMPRSS15
|
transmembrane protease, serine 15 |
| chr16_+_29690358 | 0.04 |
ENST00000395384.4
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr11_-_18343725 | 0.04 |
ENST00000531848.1
|
HPS5
|
Hermansky-Pudlak syndrome 5 |
| chr22_-_29784519 | 0.04 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr16_+_22308717 | 0.04 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr12_-_57443886 | 0.04 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr1_+_116915270 | 0.04 |
ENST00000418797.1
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chrX_+_49091920 | 0.04 |
ENST00000376227.3
|
CCDC22
|
coiled-coil domain containing 22 |
| chr15_+_86087267 | 0.04 |
ENST00000558166.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr1_-_13673511 | 0.04 |
ENST00000344998.3
ENST00000334600.6 |
PRAMEF14
|
PRAME family member 14 |
| chr2_-_170550842 | 0.04 |
ENST00000421028.1
|
CCDC173
|
coiled-coil domain containing 173 |
| chr14_+_96722152 | 0.04 |
ENST00000216629.6
|
BDKRB1
|
bradykinin receptor B1 |
| chr4_-_69434245 | 0.04 |
ENST00000317746.2
|
UGT2B17
|
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr6_-_159421198 | 0.04 |
ENST00000252655.1
ENST00000297262.3 ENST00000367069.2 |
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr7_+_80275752 | 0.04 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr4_-_150736962 | 0.04 |
ENST00000502345.1
ENST00000510975.1 ENST00000511993.1 |
RP11-526A4.1
|
RP11-526A4.1 |
| chr12_+_29376673 | 0.04 |
ENST00000547116.1
|
FAR2
|
fatty acyl CoA reductase 2 |
| chr5_+_169758393 | 0.04 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr7_+_79998864 | 0.04 |
ENST00000435819.1
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr11_+_57435219 | 0.04 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr1_+_144811943 | 0.04 |
ENST00000281815.8
|
NBPF9
|
neuroblastoma breakpoint family, member 9 |
| chr17_+_39421591 | 0.04 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr15_+_62853562 | 0.04 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr10_-_75676400 | 0.04 |
ENST00000412307.2
|
C10orf55
|
chromosome 10 open reading frame 55 |
| chr1_+_112016414 | 0.04 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr1_-_9811600 | 0.04 |
ENST00000435891.1
|
CLSTN1
|
calsyntenin 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 0.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.1 | GO:1903384 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.0 | 0.1 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.4 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.0 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:1901317 | regulation of sperm motility(GO:1901317) |
| 0.0 | 0.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.1 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.0 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.4 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.2 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |