Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
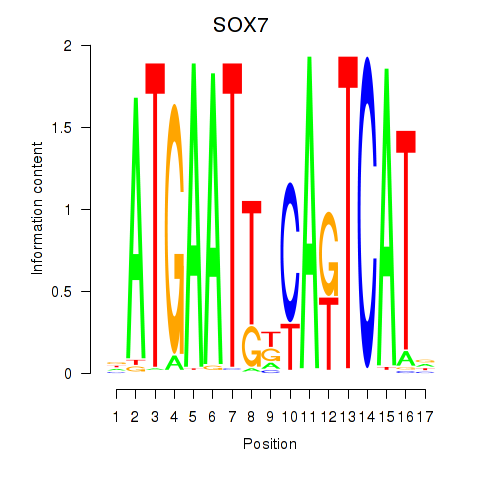
Results for SOX7
Z-value: 0.74
Transcription factors associated with SOX7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX7
|
ENSG00000171056.6 | SRY-box transcription factor 7 |
|
SOX7
|
ENSG00000258724.1 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX7 | hg19_v2_chr8_-_10588010_10588030 | -0.62 | 1.9e-01 | Click! |
Activity profile of SOX7 motif
Sorted Z-values of SOX7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_34144411 | 0.52 |
ENST00000382375.4
ENST00000453404.1 ENST00000382378.1 ENST00000477513.1 |
C21orf49
|
chromosome 21 open reading frame 49 |
| chr14_+_101361107 | 0.43 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chr15_+_59439899 | 0.39 |
ENST00000599727.1
|
C15ORF31
|
C15ORF31 |
| chr2_-_152118276 | 0.38 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr6_+_168434678 | 0.29 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr19_-_21950332 | 0.27 |
ENST00000598026.1
|
ZNF100
|
zinc finger protein 100 |
| chr1_+_10509971 | 0.25 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr16_+_22518495 | 0.25 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr12_-_57443886 | 0.23 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr19_-_38916839 | 0.23 |
ENST00000433821.2
ENST00000426920.2 ENST00000587753.1 ENST00000454404.2 ENST00000293062.9 |
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr19_-_5784610 | 0.21 |
ENST00000390672.2
ENST00000419421.2 |
PRR22
|
proline rich 22 |
| chr10_-_71169031 | 0.20 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr17_-_28661065 | 0.19 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr8_+_74271144 | 0.16 |
ENST00000519134.1
ENST00000518355.1 |
RP11-434I12.3
|
RP11-434I12.3 |
| chr6_-_111804905 | 0.16 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr5_-_95158644 | 0.16 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr15_+_75639372 | 0.15 |
ENST00000566313.1
ENST00000568059.1 ENST00000568881.1 |
NEIL1
|
nei endonuclease VIII-like 1 (E. coli) |
| chr17_-_39623681 | 0.14 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chrY_+_14813160 | 0.14 |
ENST00000338981.3
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr10_+_5005445 | 0.13 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr1_+_100598742 | 0.13 |
ENST00000370139.1
|
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr9_-_70465758 | 0.13 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr11_+_3011093 | 0.12 |
ENST00000332881.2
|
AC131971.1
|
HCG1782999; PRO0943; Uncharacterized protein |
| chr19_+_42349092 | 0.12 |
ENST00000269945.3
ENST00000596258.1 |
DMRTC2
|
DMRT-like family C2 |
| chr10_-_17243579 | 0.11 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr4_-_47983519 | 0.10 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr2_-_190445499 | 0.10 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr10_+_24738355 | 0.10 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr1_-_43751230 | 0.10 |
ENST00000523677.1
|
C1orf210
|
chromosome 1 open reading frame 210 |
| chr6_+_42531798 | 0.10 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr5_-_95158375 | 0.10 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr18_+_32820990 | 0.09 |
ENST00000601719.1
ENST00000591206.1 ENST00000330501.7 ENST00000261333.6 ENST00000355632.4 ENST00000585800.1 |
ZNF397
|
zinc finger protein 397 |
| chr19_+_37837218 | 0.09 |
ENST00000591134.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr1_-_89738458 | 0.09 |
ENST00000443807.1
|
GBP5
|
guanylate binding protein 5 |
| chr6_+_150690028 | 0.08 |
ENST00000229447.5
ENST00000344419.3 |
IYD
|
iodotyrosine deiodinase |
| chr12_-_57328187 | 0.08 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr1_+_156698743 | 0.08 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr3_+_107318157 | 0.08 |
ENST00000406780.1
|
BBX
|
bobby sox homolog (Drosophila) |
| chr10_+_122610687 | 0.07 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr1_+_158901329 | 0.07 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr12_-_56693758 | 0.07 |
ENST00000547298.1
ENST00000551936.1 ENST00000551253.1 ENST00000551473.1 |
CS
|
citrate synthase |
| chr16_+_28303804 | 0.07 |
ENST00000341901.4
|
SBK1
|
SH3 domain binding kinase 1 |
| chr14_+_50291993 | 0.07 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr1_+_223101757 | 0.07 |
ENST00000284476.6
|
DISP1
|
dispatched homolog 1 (Drosophila) |
| chr16_-_28303360 | 0.07 |
ENST00000501520.1
|
RP11-57A19.2
|
RP11-57A19.2 |
| chr9_+_105757590 | 0.07 |
ENST00000374798.3
ENST00000487798.1 |
CYLC2
|
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr1_+_100598691 | 0.06 |
ENST00000370143.1
ENST00000370141.2 |
TRMT13
|
tRNA methyltransferase 13 homolog (S. cerevisiae) |
| chr19_-_38916822 | 0.06 |
ENST00000586305.1
|
RASGRP4
|
RAS guanyl releasing protein 4 |
| chr2_+_179317994 | 0.06 |
ENST00000375129.4
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr10_-_12084770 | 0.05 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr6_+_131894284 | 0.05 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr1_+_156698708 | 0.05 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr10_-_61495760 | 0.05 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr5_-_176778803 | 0.04 |
ENST00000303127.7
|
LMAN2
|
lectin, mannose-binding 2 |
| chr14_-_31856397 | 0.03 |
ENST00000538864.2
ENST00000550366.1 |
HEATR5A
|
HEAT repeat containing 5A |
| chr5_+_175511859 | 0.03 |
ENST00000503724.2
ENST00000253490.4 |
FAM153B
|
family with sequence similarity 153, member B |
| chr10_-_5046042 | 0.03 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr7_-_143966381 | 0.03 |
ENST00000487179.1
|
CTAGE8
|
CTAGE family, member 8 |
| chr16_-_21442874 | 0.02 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr13_-_37633567 | 0.02 |
ENST00000464744.1
|
SUPT20H
|
suppressor of Ty 20 homolog (S. cerevisiae) |
| chr3_-_161089289 | 0.02 |
ENST00000497137.1
|
SPTSSB
|
serine palmitoyltransferase, small subunit B |
| chr8_+_77593448 | 0.02 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr7_-_33148975 | 0.02 |
ENST00000297157.3
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr1_-_54303934 | 0.01 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr12_+_64173583 | 0.01 |
ENST00000261234.6
|
TMEM5
|
transmembrane protein 5 |
| chr10_+_96162242 | 0.01 |
ENST00000225235.4
|
TBC1D12
|
TBC1 domain family, member 12 |
| chr8_-_82359662 | 0.01 |
ENST00000519260.1
ENST00000256103.2 |
PMP2
|
peripheral myelin protein 2 |
| chrX_+_69353284 | 0.01 |
ENST00000342206.6
ENST00000356413.4 |
IGBP1
|
immunoglobulin (CD79A) binding protein 1 |
| chr2_-_88285309 | 0.01 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr8_+_12803176 | 0.00 |
ENST00000524591.2
|
KIAA1456
|
KIAA1456 |
| chr2_+_87144738 | 0.00 |
ENST00000559485.1
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr9_-_37592561 | 0.00 |
ENST00000544379.1
ENST00000377773.5 ENST00000401811.3 ENST00000321301.6 |
TOMM5
|
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr11_+_7618413 | 0.00 |
ENST00000528883.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.2 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.1 | GO:0060345 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.0 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.1 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.0 | 0.2 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.1 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |