Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
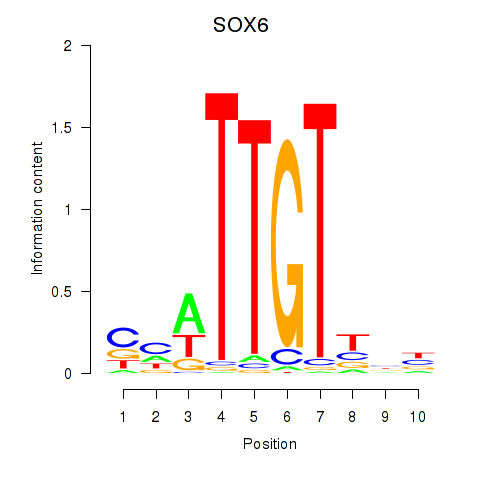
Results for SOX6
Z-value: 0.36
Transcription factors associated with SOX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX6
|
ENSG00000110693.11 | SRY-box transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX6 | hg19_v2_chr11_-_16430399_16430440 | -0.06 | 9.1e-01 | Click! |
Activity profile of SOX6 motif
Sorted Z-values of SOX6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_58145889 | 0.38 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr8_-_11660077 | 0.23 |
ENST00000533405.1
|
RP11-297N6.4
|
Uncharacterized protein |
| chr5_-_39424961 | 0.18 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_+_23146271 | 0.18 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr15_-_80695917 | 0.17 |
ENST00000559008.1
|
RP11-210M15.2
|
Uncharacterized protein |
| chr13_+_24144796 | 0.16 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr19_-_18717627 | 0.16 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr6_-_108278456 | 0.15 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr17_+_1666108 | 0.15 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr20_+_34679725 | 0.14 |
ENST00000432589.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr3_-_8686479 | 0.14 |
ENST00000544814.1
ENST00000427408.1 |
SSUH2
|
ssu-2 homolog (C. elegans) |
| chr2_-_86564776 | 0.13 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr12_+_124997766 | 0.13 |
ENST00000543970.1
|
RP11-83B20.1
|
RP11-83B20.1 |
| chr5_+_140254884 | 0.12 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr5_-_146833803 | 0.12 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr3_-_197024394 | 0.11 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr17_+_76037081 | 0.11 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr13_+_24144509 | 0.11 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr12_-_11036844 | 0.11 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr3_-_15563229 | 0.10 |
ENST00000383786.5
ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr10_+_22605374 | 0.10 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chr16_+_23765948 | 0.10 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr2_-_114647327 | 0.10 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chrX_+_12993202 | 0.10 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr8_+_107738240 | 0.10 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr18_-_73967160 | 0.10 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr4_+_70916119 | 0.10 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr7_-_50628745 | 0.10 |
ENST00000380984.4
ENST00000357936.5 ENST00000426377.1 |
DDC
|
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr2_+_42104692 | 0.10 |
ENST00000398796.2
ENST00000442214.1 |
AC104654.1
|
AC104654.1 |
| chr11_+_19799327 | 0.09 |
ENST00000540292.1
|
NAV2
|
neuron navigator 2 |
| chr7_+_69064566 | 0.09 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr19_-_17571722 | 0.09 |
ENST00000301944.2
|
NXNL1
|
nucleoredoxin-like 1 |
| chr3_+_183353356 | 0.09 |
ENST00000242810.6
ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24
|
kelch-like family member 24 |
| chr11_+_125365110 | 0.09 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chr1_-_65533390 | 0.09 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr20_+_48884002 | 0.09 |
ENST00000425497.1
ENST00000445003.1 |
RP11-290F20.3
|
RP11-290F20.3 |
| chr16_-_15736881 | 0.09 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr9_-_130712995 | 0.09 |
ENST00000373084.4
|
FAM102A
|
family with sequence similarity 102, member A |
| chr22_-_42336209 | 0.09 |
ENST00000472374.2
|
CENPM
|
centromere protein M |
| chr11_+_19798964 | 0.09 |
ENST00000527559.2
|
NAV2
|
neuron navigator 2 |
| chr2_-_47572105 | 0.08 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr6_-_127840048 | 0.08 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr14_+_20215587 | 0.08 |
ENST00000331723.1
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3 |
| chr9_-_98268883 | 0.08 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr2_-_86564740 | 0.08 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
| chr1_+_14075903 | 0.08 |
ENST00000343137.4
ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr13_+_110958124 | 0.08 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr12_-_48213735 | 0.07 |
ENST00000417902.1
ENST00000417107.1 |
HDAC7
|
histone deacetylase 7 |
| chr1_-_204436344 | 0.07 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr2_+_30569506 | 0.07 |
ENST00000421976.2
|
AC109642.1
|
AC109642.1 |
| chr7_+_69064300 | 0.07 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr6_+_13272904 | 0.07 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr6_-_42110342 | 0.07 |
ENST00000356542.5
|
C6orf132
|
chromosome 6 open reading frame 132 |
| chr6_-_139613269 | 0.07 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr17_-_39041479 | 0.07 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr1_+_1361494 | 0.07 |
ENST00000378821.3
|
TMEM88B
|
transmembrane protein 88B |
| chrX_+_80457442 | 0.07 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chrX_+_84498989 | 0.07 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chrX_+_12993336 | 0.07 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr11_-_130184470 | 0.06 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr8_+_107738343 | 0.06 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chr12_-_109797249 | 0.06 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr16_+_56970567 | 0.06 |
ENST00000563911.1
|
HERPUD1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr16_+_72088376 | 0.06 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr13_-_24007815 | 0.06 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr16_-_3068171 | 0.06 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr3_+_151986709 | 0.06 |
ENST00000495875.2
ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1
|
muscleblind-like splicing regulator 1 |
| chr14_+_74815116 | 0.06 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated |
| chr22_+_43808014 | 0.06 |
ENST00000334209.5
ENST00000443721.1 ENST00000414469.2 ENST00000439548.1 |
MPPED1
|
metallophosphoesterase domain containing 1 |
| chr10_-_25241499 | 0.06 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr12_+_27623565 | 0.06 |
ENST00000535986.1
|
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr8_-_102218292 | 0.06 |
ENST00000518336.1
ENST00000520454.1 |
ZNF706
|
zinc finger protein 706 |
| chr15_+_52043813 | 0.06 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chr4_+_78078304 | 0.06 |
ENST00000316355.5
ENST00000354403.5 ENST00000502280.1 |
CCNG2
|
cyclin G2 |
| chr2_-_55276320 | 0.06 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr4_+_2819883 | 0.06 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chrX_+_84499038 | 0.06 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr3_+_186649133 | 0.06 |
ENST00000417392.1
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr3_-_57583052 | 0.06 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr6_-_35464817 | 0.06 |
ENST00000338863.7
|
TEAD3
|
TEA domain family member 3 |
| chr1_+_228337553 | 0.06 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr6_+_122720681 | 0.06 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr15_+_52043758 | 0.05 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr1_-_6479963 | 0.05 |
ENST00000377836.4
ENST00000487437.1 ENST00000489730.1 ENST00000377834.4 |
HES2
|
hes family bHLH transcription factor 2 |
| chr12_-_58145604 | 0.05 |
ENST00000552254.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr4_-_105416039 | 0.05 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr22_+_18593097 | 0.05 |
ENST00000426208.1
|
TUBA8
|
tubulin, alpha 8 |
| chr12_+_9066472 | 0.05 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr7_-_82792215 | 0.05 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr9_-_134145880 | 0.05 |
ENST00000372269.3
ENST00000464831.1 |
FAM78A
|
family with sequence similarity 78, member A |
| chr16_-_67970990 | 0.05 |
ENST00000358514.4
|
PSMB10
|
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr19_-_44124019 | 0.05 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chr16_-_4852915 | 0.05 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr10_+_24528108 | 0.05 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr4_+_159690218 | 0.05 |
ENST00000264433.6
|
FNIP2
|
folliculin interacting protein 2 |
| chr11_-_82708435 | 0.05 |
ENST00000525117.1
ENST00000532548.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chrX_-_45060135 | 0.05 |
ENST00000398000.2
ENST00000377934.4 |
CXorf36
|
chromosome X open reading frame 36 |
| chr1_+_244998602 | 0.05 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr11_-_130184555 | 0.05 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr4_+_88896819 | 0.05 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr22_-_22292934 | 0.05 |
ENST00000538191.1
ENST00000424647.1 ENST00000407142.1 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr5_-_157002749 | 0.04 |
ENST00000517905.1
ENST00000430702.2 ENST00000394020.1 |
ADAM19
|
ADAM metallopeptidase domain 19 |
| chr1_+_202183200 | 0.04 |
ENST00000439764.2
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr8_+_27631903 | 0.04 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr6_-_133055815 | 0.04 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr7_+_139528952 | 0.04 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr15_+_89631647 | 0.04 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr20_-_39317868 | 0.04 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr4_+_26585538 | 0.04 |
ENST00000264866.4
|
TBC1D19
|
TBC1 domain family, member 19 |
| chr6_-_35464727 | 0.04 |
ENST00000402886.3
|
TEAD3
|
TEA domain family member 3 |
| chr20_-_35492048 | 0.04 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr8_-_42752418 | 0.04 |
ENST00000524954.1
|
RNF170
|
ring finger protein 170 |
| chr2_-_208890218 | 0.04 |
ENST00000457206.1
ENST00000427836.2 ENST00000389247.4 |
PLEKHM3
|
pleckstrin homology domain containing, family M, member 3 |
| chr12_-_49582593 | 0.04 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr11_+_36317830 | 0.04 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr8_-_42358742 | 0.04 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr2_-_42180940 | 0.04 |
ENST00000378711.2
|
C2orf91
|
chromosome 2 open reading frame 91 |
| chr1_-_161014731 | 0.04 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr17_-_53046058 | 0.04 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr15_-_82338460 | 0.04 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr15_+_93426514 | 0.04 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr20_+_60718785 | 0.04 |
ENST00000421564.1
ENST00000450482.1 ENST00000331758.3 |
SS18L1
|
synovial sarcoma translocation gene on chromosome 18-like 1 |
| chr6_+_32936942 | 0.04 |
ENST00000496118.2
|
BRD2
|
bromodomain containing 2 |
| chr14_-_96830207 | 0.04 |
ENST00000359933.4
|
ATG2B
|
autophagy related 2B |
| chr2_-_39348137 | 0.04 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr4_-_186732048 | 0.04 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr4_+_89300158 | 0.04 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr16_+_3704822 | 0.04 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr13_+_100153665 | 0.03 |
ENST00000376387.4
|
TM9SF2
|
transmembrane 9 superfamily member 2 |
| chr1_+_9005917 | 0.03 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chr21_+_30671690 | 0.03 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr15_+_89631381 | 0.03 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr15_-_88799948 | 0.03 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr18_-_32924372 | 0.03 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr11_-_75236867 | 0.03 |
ENST00000376282.3
ENST00000336898.3 |
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr4_+_95129061 | 0.03 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr15_+_85923797 | 0.03 |
ENST00000559362.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr1_+_25071848 | 0.03 |
ENST00000374379.4
|
CLIC4
|
chloride intracellular channel 4 |
| chr2_-_166060552 | 0.03 |
ENST00000283254.7
ENST00000453007.1 |
SCN3A
|
sodium channel, voltage-gated, type III, alpha subunit |
| chrX_+_107288239 | 0.03 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr17_-_49124230 | 0.03 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr1_+_196743912 | 0.03 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr6_-_127840336 | 0.03 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr4_+_26585686 | 0.03 |
ENST00000505206.1
ENST00000511789.1 |
TBC1D19
|
TBC1 domain family, member 19 |
| chr1_+_24286287 | 0.03 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr3_-_57583130 | 0.03 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr8_+_24151553 | 0.03 |
ENST00000265769.4
ENST00000540823.1 ENST00000397649.3 |
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr14_-_91884115 | 0.03 |
ENST00000389857.6
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr14_-_71107921 | 0.03 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr12_-_49582978 | 0.03 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr6_-_132834184 | 0.03 |
ENST00000367941.2
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr10_-_13390270 | 0.03 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr9_-_98269699 | 0.03 |
ENST00000429896.2
|
PTCH1
|
patched 1 |
| chr15_+_81293254 | 0.03 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr17_+_64961026 | 0.03 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr11_+_64949899 | 0.03 |
ENST00000531068.1
ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr22_-_21984282 | 0.03 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr6_-_29527702 | 0.03 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr6_-_128841503 | 0.03 |
ENST00000368215.3
ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK
|
protein tyrosine phosphatase, receptor type, K |
| chr12_+_22778009 | 0.03 |
ENST00000266517.4
ENST00000335148.3 |
ETNK1
|
ethanolamine kinase 1 |
| chr20_-_17511962 | 0.03 |
ENST00000377873.3
|
BFSP1
|
beaded filament structural protein 1, filensin |
| chr19_-_17571613 | 0.03 |
ENST00000594663.1
|
CTD-2521M24.10
|
Uncharacterized protein |
| chr17_+_53046096 | 0.03 |
ENST00000376352.2
ENST00000299341.4 ENST00000405898.1 ENST00000434978.2 ENST00000398391.2 |
STXBP4
|
syntaxin binding protein 4 |
| chr1_+_150229554 | 0.03 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr5_+_86563636 | 0.03 |
ENST00000274376.6
|
RASA1
|
RAS p21 protein activator (GTPase activating protein) 1 |
| chr6_+_32937083 | 0.03 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr16_+_68573640 | 0.03 |
ENST00000398253.2
ENST00000573161.1 |
ZFP90
|
ZFP90 zinc finger protein |
| chr2_-_157198860 | 0.03 |
ENST00000409572.1
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr17_+_72667239 | 0.03 |
ENST00000402449.4
|
RAB37
|
RAB37, member RAS oncogene family |
| chr2_+_65215604 | 0.02 |
ENST00000531327.1
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr5_+_139175380 | 0.02 |
ENST00000274710.3
|
PSD2
|
pleckstrin and Sec7 domain containing 2 |
| chr11_-_64013663 | 0.02 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr3_+_72201910 | 0.02 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr1_-_32801825 | 0.02 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr9_-_98269481 | 0.02 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr4_-_69111401 | 0.02 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr12_-_99288986 | 0.02 |
ENST00000552407.1
ENST00000551613.1 ENST00000548447.1 ENST00000546364.3 ENST00000552748.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr19_-_59030921 | 0.02 |
ENST00000354590.3
ENST00000596739.1 |
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr16_+_3162557 | 0.02 |
ENST00000382192.3
ENST00000219091.4 ENST00000444510.2 ENST00000414351.1 |
ZNF205
|
zinc finger protein 205 |
| chr1_-_1243392 | 0.02 |
ENST00000354700.5
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr3_-_167813672 | 0.02 |
ENST00000470487.1
|
GOLIM4
|
golgi integral membrane protein 4 |
| chr4_-_87028478 | 0.02 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr2_-_70475730 | 0.02 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr17_-_46716647 | 0.02 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr5_-_76788317 | 0.02 |
ENST00000296679.4
|
WDR41
|
WD repeat domain 41 |
| chr19_+_49468558 | 0.02 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr20_+_6748311 | 0.02 |
ENST00000378827.4
|
BMP2
|
bone morphogenetic protein 2 |
| chr4_+_95128748 | 0.02 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr12_-_58146048 | 0.02 |
ENST00000547281.1
ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4
|
cyclin-dependent kinase 4 |
| chr1_-_155880672 | 0.02 |
ENST00000609492.1
ENST00000368322.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr2_-_47572207 | 0.02 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr4_-_52883786 | 0.02 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr4_-_140477928 | 0.02 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr7_+_29234101 | 0.02 |
ENST00000435288.2
|
CHN2
|
chimerin 2 |
| chr9_-_123476719 | 0.02 |
ENST00000373930.3
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr15_+_74610894 | 0.02 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr15_+_52311398 | 0.02 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr12_+_25205666 | 0.02 |
ENST00000547044.1
|
LRMP
|
lymphoid-restricted membrane protein |
| chr1_-_26232522 | 0.02 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr5_-_76788134 | 0.02 |
ENST00000507029.1
|
WDR41
|
WD repeat domain 41 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX6
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.0 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 | 0.1 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.2 | GO:0021564 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.0 | GO:0035284 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.0 | GO:2000866 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.0 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |