Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
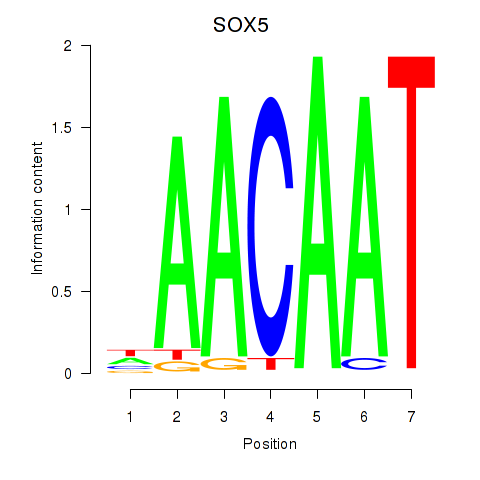
Results for SOX5
Z-value: 0.89
Transcription factors associated with SOX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX5
|
ENSG00000134532.11 | SRY-box transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX5 | hg19_v2_chr12_-_24103954_24103972 | 0.41 | 4.2e-01 | Click! |
Activity profile of SOX5 motif
Sorted Z-values of SOX5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_98268883 | 0.68 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr2_-_25451065 | 0.49 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr16_+_30934376 | 0.48 |
ENST00000562798.1
ENST00000471231.2 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr14_-_92414294 | 0.44 |
ENST00000554468.1
|
FBLN5
|
fibulin 5 |
| chr1_-_40367668 | 0.38 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr14_-_92414055 | 0.36 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr8_-_128231299 | 0.32 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr6_+_12290586 | 0.31 |
ENST00000379375.5
|
EDN1
|
endothelin 1 |
| chr5_-_138780159 | 0.29 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr1_+_43637996 | 0.27 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr2_-_145188137 | 0.27 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr11_-_125365435 | 0.27 |
ENST00000524435.1
|
FEZ1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr17_+_40119801 | 0.26 |
ENST00000585452.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr3_+_189507460 | 0.25 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr1_-_161008697 | 0.25 |
ENST00000318289.10
ENST00000368023.3 ENST00000368024.1 ENST00000423014.2 |
TSTD1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr14_-_38036271 | 0.24 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr14_-_92413727 | 0.23 |
ENST00000267620.10
|
FBLN5
|
fibulin 5 |
| chr14_-_105071083 | 0.22 |
ENST00000415614.2
|
TMEM179
|
transmembrane protein 179 |
| chr12_-_71003568 | 0.21 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr12_+_78359999 | 0.21 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr6_+_105404899 | 0.21 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chr10_+_35415851 | 0.21 |
ENST00000374726.3
|
CREM
|
cAMP responsive element modulator |
| chr2_+_27070964 | 0.21 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr3_+_181429704 | 0.21 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr20_+_34679725 | 0.20 |
ENST00000432589.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr4_+_76439095 | 0.20 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr14_+_97925151 | 0.19 |
ENST00000554862.1
ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1
|
CTD-2506J14.1 |
| chr2_-_188312971 | 0.19 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr18_+_21594384 | 0.19 |
ENST00000584250.1
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr12_-_12674032 | 0.19 |
ENST00000298573.4
|
DUSP16
|
dual specificity phosphatase 16 |
| chr11_+_46402583 | 0.19 |
ENST00000359803.3
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr15_-_41166414 | 0.19 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr6_+_126240442 | 0.18 |
ENST00000448104.1
ENST00000438495.1 ENST00000444128.1 |
NCOA7
|
nuclear receptor coactivator 7 |
| chr22_-_36236265 | 0.17 |
ENST00000414461.2
ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr7_-_83824169 | 0.17 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr10_-_118897806 | 0.17 |
ENST00000277905.2
|
VAX1
|
ventral anterior homeobox 1 |
| chr3_-_114790179 | 0.16 |
ENST00000462705.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr11_+_111808119 | 0.16 |
ENST00000531396.1
|
DIXDC1
|
DIX domain containing 1 |
| chr17_+_33448593 | 0.16 |
ENST00000158009.5
|
FNDC8
|
fibronectin type III domain containing 8 |
| chr10_-_14614311 | 0.15 |
ENST00000479731.1
ENST00000468492.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr2_+_110656268 | 0.15 |
ENST00000553749.1
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr4_-_184241927 | 0.15 |
ENST00000323319.5
|
CLDN22
|
claudin 22 |
| chr20_-_45985464 | 0.15 |
ENST00000458360.2
ENST00000262975.4 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr16_+_86612112 | 0.15 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr14_-_89960395 | 0.15 |
ENST00000555034.1
ENST00000553904.1 |
FOXN3
|
forkhead box N3 |
| chr6_+_21666633 | 0.14 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chrX_+_135251783 | 0.14 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr2_-_192711968 | 0.14 |
ENST00000304141.4
|
SDPR
|
serum deprivation response |
| chr3_+_112930306 | 0.14 |
ENST00000495514.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr2_+_27071045 | 0.14 |
ENST00000401478.1
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr17_+_30814707 | 0.14 |
ENST00000584792.1
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr12_+_49761147 | 0.14 |
ENST00000549298.1
|
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr17_+_67498295 | 0.13 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr17_+_1666108 | 0.13 |
ENST00000570731.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr14_-_69619823 | 0.13 |
ENST00000341516.5
|
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr20_-_45985172 | 0.13 |
ENST00000536340.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr12_-_96793142 | 0.13 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr9_+_129097479 | 0.13 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B |
| chr18_+_3451646 | 0.13 |
ENST00000345133.5
ENST00000330513.5 ENST00000549546.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_+_112929850 | 0.13 |
ENST00000464546.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chrY_+_15016013 | 0.13 |
ENST00000360160.4
ENST00000454054.1 |
DDX3Y
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr10_-_14613968 | 0.13 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr11_+_110001723 | 0.12 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr9_+_118916082 | 0.12 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chr8_-_95449155 | 0.12 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr10_-_104913367 | 0.12 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr10_-_14614122 | 0.12 |
ENST00000378465.3
ENST00000452706.2 ENST00000378458.2 |
FAM107B
|
family with sequence similarity 107, member B |
| chr7_-_83824449 | 0.12 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr7_+_134576317 | 0.12 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr7_-_7575477 | 0.12 |
ENST00000399429.3
|
COL28A1
|
collagen, type XXVIII, alpha 1 |
| chr7_-_27142290 | 0.12 |
ENST00000222718.5
|
HOXA2
|
homeobox A2 |
| chr18_+_3451584 | 0.11 |
ENST00000551541.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr12_-_52715179 | 0.11 |
ENST00000293670.3
|
KRT83
|
keratin 83 |
| chr17_+_67498396 | 0.11 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr17_-_47786375 | 0.11 |
ENST00000511657.1
|
SLC35B1
|
solute carrier family 35, member B1 |
| chr3_+_189507523 | 0.11 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr2_+_33661382 | 0.11 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chrX_+_135252050 | 0.11 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr10_-_14614095 | 0.11 |
ENST00000482277.1
ENST00000378462.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chrX_+_70503433 | 0.10 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr7_-_47621736 | 0.10 |
ENST00000311160.9
|
TNS3
|
tensin 3 |
| chr10_+_35416090 | 0.10 |
ENST00000354759.3
|
CREM
|
cAMP responsive element modulator |
| chr5_-_39462390 | 0.10 |
ENST00000511792.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr12_+_498545 | 0.10 |
ENST00000543504.1
|
CCDC77
|
coiled-coil domain containing 77 |
| chr12_+_498500 | 0.10 |
ENST00000540180.1
ENST00000422000.1 ENST00000535052.1 |
CCDC77
|
coiled-coil domain containing 77 |
| chr7_+_129015671 | 0.10 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr8_-_72274467 | 0.10 |
ENST00000340726.3
|
EYA1
|
eyes absent homolog 1 (Drosophila) |
| chr3_-_123339343 | 0.10 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr5_+_179159813 | 0.10 |
ENST00000292599.3
|
MAML1
|
mastermind-like 1 (Drosophila) |
| chr12_+_9066472 | 0.10 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr14_-_92413353 | 0.10 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr1_-_11751529 | 0.09 |
ENST00000376672.1
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr8_-_40755333 | 0.09 |
ENST00000297737.6
ENST00000315769.7 |
ZMAT4
|
zinc finger, matrin-type 4 |
| chr1_-_11751665 | 0.09 |
ENST00000376667.3
ENST00000235310.3 |
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast) |
| chr6_+_89790459 | 0.09 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr11_+_46402744 | 0.09 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chrX_+_135251835 | 0.09 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr12_-_122985494 | 0.09 |
ENST00000336229.4
|
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr19_-_7698599 | 0.09 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr1_-_232651312 | 0.09 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr9_+_129097520 | 0.09 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr4_+_71588372 | 0.09 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr20_-_45985414 | 0.09 |
ENST00000461685.1
ENST00000372023.3 ENST00000540497.1 ENST00000435836.1 ENST00000471951.2 ENST00000352431.2 ENST00000396281.4 ENST00000355972.4 ENST00000360911.3 |
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr16_+_3704822 | 0.09 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr14_-_69619291 | 0.09 |
ENST00000554215.1
ENST00000556847.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr14_+_61654271 | 0.09 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr15_-_86338100 | 0.09 |
ENST00000536947.1
|
KLHL25
|
kelch-like family member 25 |
| chr17_-_39041479 | 0.09 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr4_-_83350580 | 0.09 |
ENST00000349655.4
ENST00000602300.1 |
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr2_-_166702601 | 0.09 |
ENST00000428888.1
|
AC009495.4
|
AC009495.4 |
| chr11_-_82708435 | 0.08 |
ENST00000525117.1
ENST00000532548.1 |
RAB30
|
RAB30, member RAS oncogene family |
| chr7_-_47622156 | 0.08 |
ENST00000457718.1
|
TNS3
|
tensin 3 |
| chr7_+_77167343 | 0.08 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr2_+_232573222 | 0.08 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr3_-_99594948 | 0.08 |
ENST00000471562.1
ENST00000495625.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr3_-_19988462 | 0.08 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr8_+_26435915 | 0.08 |
ENST00000523027.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr2_+_102314161 | 0.08 |
ENST00000425019.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr7_+_77167376 | 0.08 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr12_+_32655110 | 0.08 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr12_+_50479109 | 0.08 |
ENST00000550477.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr6_-_99873145 | 0.08 |
ENST00000369239.5
ENST00000438806.1 |
PNISR
|
PNN-interacting serine/arginine-rich protein |
| chr15_+_57211318 | 0.08 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr1_+_147013182 | 0.08 |
ENST00000234739.3
|
BCL9
|
B-cell CLL/lymphoma 9 |
| chr4_-_83351005 | 0.08 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr3_-_141747459 | 0.08 |
ENST00000477292.1
ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chrX_-_13835147 | 0.08 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr11_-_115375107 | 0.08 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chrX_-_107682702 | 0.08 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr4_+_165675269 | 0.07 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr13_+_73629107 | 0.07 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr15_+_84115868 | 0.07 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr13_+_103459704 | 0.07 |
ENST00000602836.1
|
BIVM-ERCC5
|
BIVM-ERCC5 readthrough |
| chr4_-_69215699 | 0.07 |
ENST00000510746.1
ENST00000344157.4 ENST00000355665.3 |
YTHDC1
|
YTH domain containing 1 |
| chrX_-_74145273 | 0.07 |
ENST00000055682.6
|
KIAA2022
|
KIAA2022 |
| chr9_-_15510989 | 0.07 |
ENST00000380715.1
ENST00000380716.4 ENST00000380738.4 ENST00000380733.4 |
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr12_-_122985067 | 0.07 |
ENST00000540586.1
ENST00000543897.1 |
ZCCHC8
|
zinc finger, CCHC domain containing 8 |
| chr11_+_57435219 | 0.07 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr11_+_111807863 | 0.07 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr11_+_36397915 | 0.07 |
ENST00000526682.1
ENST00000530252.1 |
PRR5L
|
proline rich 5 like |
| chr5_+_138678131 | 0.07 |
ENST00000394795.2
ENST00000510080.1 |
PAIP2
|
poly(A) binding protein interacting protein 2 |
| chr3_+_19988736 | 0.07 |
ENST00000443878.1
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr12_+_49761224 | 0.07 |
ENST00000553127.1
ENST00000321898.6 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr2_+_232573208 | 0.07 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr14_-_73360796 | 0.07 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr6_-_79944336 | 0.07 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr3_+_189349162 | 0.07 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr14_-_69619689 | 0.06 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr18_-_52989217 | 0.06 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr17_-_56082455 | 0.06 |
ENST00000578794.1
|
RP11-159D12.5
|
Uncharacterized protein |
| chrX_+_107683096 | 0.06 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr12_+_56360605 | 0.06 |
ENST00000553376.1
ENST00000440311.2 ENST00000354056.4 |
CDK2
|
cyclin-dependent kinase 2 |
| chr10_-_104953009 | 0.06 |
ENST00000470299.1
ENST00000343289.5 |
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr6_-_42110342 | 0.06 |
ENST00000356542.5
|
C6orf132
|
chromosome 6 open reading frame 132 |
| chr12_+_49761273 | 0.06 |
ENST00000551540.1
ENST00000552918.1 ENST00000548777.1 ENST00000547865.1 ENST00000552171.1 |
SPATS2
|
spermatogenesis associated, serine-rich 2 |
| chr6_+_88054530 | 0.06 |
ENST00000388923.4
|
C6orf163
|
chromosome 6 open reading frame 163 |
| chr10_+_104535994 | 0.06 |
ENST00000369889.4
|
WBP1L
|
WW domain binding protein 1-like |
| chr3_-_129407535 | 0.06 |
ENST00000432054.2
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr20_-_52612705 | 0.06 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr1_+_165797024 | 0.06 |
ENST00000372212.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_+_164529004 | 0.06 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr17_-_4269920 | 0.06 |
ENST00000572484.1
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr3_+_120315160 | 0.06 |
ENST00000485064.1
ENST00000492739.1 |
NDUFB4
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
| chr20_-_45981138 | 0.06 |
ENST00000446994.2
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr17_+_37894179 | 0.06 |
ENST00000577695.1
ENST00000309156.4 ENST00000309185.3 |
GRB7
|
growth factor receptor-bound protein 7 |
| chrX_+_70503037 | 0.06 |
ENST00000535149.1
|
NONO
|
non-POU domain containing, octamer-binding |
| chr17_-_73505961 | 0.05 |
ENST00000433559.2
|
CASKIN2
|
CASK interacting protein 2 |
| chrX_+_9880412 | 0.05 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr15_+_90319557 | 0.05 |
ENST00000341735.3
|
MESP2
|
mesoderm posterior 2 homolog (mouse) |
| chrX_+_78003204 | 0.05 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr18_-_53177984 | 0.05 |
ENST00000543082.1
|
TCF4
|
transcription factor 4 |
| chr5_+_140235469 | 0.05 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr12_-_111358372 | 0.05 |
ENST00000548438.1
ENST00000228841.8 |
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow |
| chr11_-_72852958 | 0.05 |
ENST00000458644.2
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chrX_-_142722897 | 0.05 |
ENST00000338017.4
|
SLITRK4
|
SLIT and NTRK-like family, member 4 |
| chr10_+_35416223 | 0.05 |
ENST00000489321.1
ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM
|
cAMP responsive element modulator |
| chr6_-_9939552 | 0.05 |
ENST00000460363.2
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr8_-_116673894 | 0.05 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr1_-_36906474 | 0.05 |
ENST00000433045.2
|
OSCP1
|
organic solute carrier partner 1 |
| chr7_+_134551583 | 0.05 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr12_+_50479101 | 0.05 |
ENST00000551966.1
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr11_-_64647144 | 0.05 |
ENST00000359393.2
ENST00000433803.1 ENST00000411683.1 |
EHD1
|
EH-domain containing 1 |
| chr1_-_23340400 | 0.05 |
ENST00000440767.2
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr10_+_52751010 | 0.05 |
ENST00000373985.1
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr10_+_63661053 | 0.05 |
ENST00000279873.7
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like) |
| chr10_+_70480963 | 0.05 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr11_+_73359719 | 0.05 |
ENST00000535129.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr22_+_43808014 | 0.05 |
ENST00000334209.5
ENST00000443721.1 ENST00000414469.2 ENST00000439548.1 |
MPPED1
|
metallophosphoesterase domain containing 1 |
| chr11_-_102323489 | 0.05 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr2_-_208030886 | 0.05 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr15_-_86338134 | 0.05 |
ENST00000337975.5
|
KLHL25
|
kelch-like family member 25 |
| chr12_-_110937351 | 0.05 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr4_+_41614720 | 0.05 |
ENST00000509277.1
|
LIMCH1
|
LIM and calponin homology domains 1 |
| chr18_+_21594585 | 0.05 |
ENST00000317571.3
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr8_-_30706608 | 0.05 |
ENST00000256246.2
|
TEX15
|
testis expressed 15 |
| chr15_+_66994561 | 0.04 |
ENST00000288840.5
|
SMAD6
|
SMAD family member 6 |
| chr3_-_65583561 | 0.04 |
ENST00000460329.2
|
MAGI1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr3_-_48754599 | 0.04 |
ENST00000413654.1
ENST00000454335.1 ENST00000440424.1 ENST00000449610.1 ENST00000443964.1 ENST00000417896.1 ENST00000413298.1 ENST00000449563.1 ENST00000443853.1 ENST00000437427.1 ENST00000446860.1 ENST00000412850.1 ENST00000424035.1 ENST00000340879.4 ENST00000431721.2 ENST00000434860.1 ENST00000328631.5 ENST00000432678.2 |
IP6K2
|
inositol hexakisphosphate kinase 2 |
| chr4_-_101439242 | 0.04 |
ENST00000296420.4
|
EMCN
|
endomucin |
| chr1_+_26437631 | 0.04 |
ENST00000444713.1
|
PDIK1L
|
PDLIM1 interacting kinase 1 like |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX5
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.4 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.3 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 1.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.4 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.4 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.2 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.3 | GO:1902748 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.2 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:0060922 | atrioventricular node development(GO:0003162) cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.0 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.3 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 0.7 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0000806 | Y chromosome(GO:0000806) cyclin A1-CDK2 complex(GO:0097123) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 0.3 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.2 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0070698 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |