Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
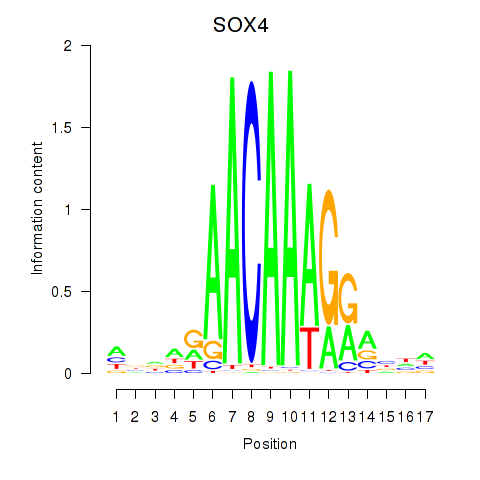
Results for SOX4
Z-value: 0.57
Transcription factors associated with SOX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX4
|
ENSG00000124766.4 | SRY-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX4 | hg19_v2_chr6_+_21593972_21594071 | 0.36 | 4.9e-01 | Click! |
Activity profile of SOX4 motif
Sorted Z-values of SOX4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_134551583 | 0.42 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr1_+_185126598 | 0.39 |
ENST00000450350.1
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr19_-_18717627 | 0.31 |
ENST00000392386.3
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr7_-_84121858 | 0.31 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_+_61473104 | 0.29 |
ENST00000583016.1
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr4_-_170192000 | 0.28 |
ENST00000502315.1
|
SH3RF1
|
SH3 domain containing ring finger 1 |
| chr15_-_74494779 | 0.27 |
ENST00000571341.1
|
STRA6
|
stimulated by retinoic acid 6 |
| chr10_-_18948208 | 0.27 |
ENST00000607346.1
|
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr1_-_204436344 | 0.26 |
ENST00000367184.2
|
PIK3C2B
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr14_-_69658127 | 0.25 |
ENST00000556182.1
|
RP11-363J20.1
|
RP11-363J20.1 |
| chr15_-_76304731 | 0.24 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr6_+_123038689 | 0.24 |
ENST00000354275.2
ENST00000368446.1 |
PKIB
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr4_-_75024085 | 0.23 |
ENST00000600169.1
|
AC093677.1
|
Uncharacterized protein |
| chr8_-_42358742 | 0.22 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr19_+_10765614 | 0.21 |
ENST00000589283.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr3_-_197024394 | 0.20 |
ENST00000434148.1
ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr12_+_8849773 | 0.17 |
ENST00000541044.1
|
RIMKLB
|
ribosomal modification protein rimK-like family member B |
| chr5_+_138852090 | 0.17 |
ENST00000593907.1
|
AC138517.1
|
Uncharacterized protein |
| chr22_-_31688431 | 0.17 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr5_+_150816595 | 0.16 |
ENST00000520111.1
ENST00000520701.1 ENST00000429484.2 |
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr19_-_38720294 | 0.16 |
ENST00000412732.1
ENST00000456296.1 |
DPF1
|
D4, zinc and double PHD fingers family 1 |
| chr12_+_70760056 | 0.16 |
ENST00000258111.4
|
KCNMB4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr14_-_51027838 | 0.15 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr13_+_98795664 | 0.15 |
ENST00000376581.5
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr16_-_31085033 | 0.14 |
ENST00000414399.1
|
ZNF668
|
zinc finger protein 668 |
| chr19_-_59030921 | 0.14 |
ENST00000354590.3
ENST00000596739.1 |
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr1_-_85930246 | 0.13 |
ENST00000426972.3
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr12_-_122296755 | 0.13 |
ENST00000289004.4
|
HPD
|
4-hydroxyphenylpyruvate dioxygenase |
| chr17_+_67498396 | 0.13 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr16_+_524900 | 0.13 |
ENST00000449879.1
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr17_+_59489112 | 0.12 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chrX_-_73513307 | 0.12 |
ENST00000602420.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr5_-_111091948 | 0.12 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr2_+_105050794 | 0.12 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr22_-_42336209 | 0.12 |
ENST00000472374.2
|
CENPM
|
centromere protein M |
| chr2_-_127977654 | 0.12 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr14_-_61124977 | 0.11 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr5_+_140235469 | 0.11 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr14_-_73493825 | 0.11 |
ENST00000318876.5
ENST00000556143.1 |
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr19_+_10765699 | 0.11 |
ENST00000590009.1
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chr14_+_24439148 | 0.11 |
ENST00000543805.1
ENST00000534993.1 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr10_-_18948156 | 0.11 |
ENST00000414939.1
ENST00000449529.1 ENST00000456217.1 ENST00000444660.1 |
ARL5B-AS1
|
ARL5B antisense RNA 1 |
| chr3_-_134092561 | 0.10 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr7_-_111032971 | 0.10 |
ENST00000450877.1
|
IMMP2L
|
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr10_-_105845674 | 0.10 |
ENST00000353479.5
ENST00000369733.3 |
COL17A1
|
collagen, type XVII, alpha 1 |
| chr22_-_21984282 | 0.10 |
ENST00000398873.3
ENST00000292778.6 |
YDJC
|
YdjC homolog (bacterial) |
| chr13_-_110959478 | 0.10 |
ENST00000543140.1
ENST00000375820.4 |
COL4A1
|
collagen, type IV, alpha 1 |
| chr14_+_101299520 | 0.10 |
ENST00000455531.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr1_+_100817262 | 0.10 |
ENST00000455467.1
|
CDC14A
|
cell division cycle 14A |
| chr2_+_220309379 | 0.09 |
ENST00000451076.1
|
SPEG
|
SPEG complex locus |
| chr22_-_31688381 | 0.09 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr4_-_140544386 | 0.09 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr8_-_9762871 | 0.09 |
ENST00000521242.1
|
LINC00599
|
long intergenic non-protein coding RNA 599 |
| chr14_+_32798547 | 0.09 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr4_-_40516560 | 0.09 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr16_+_72088376 | 0.09 |
ENST00000570083.1
ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP
HPR
|
haptoglobin haptoglobin-related protein |
| chr1_-_32801825 | 0.08 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr7_+_23146271 | 0.08 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr16_-_48281444 | 0.08 |
ENST00000537808.1
ENST00000569991.1 |
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr19_-_59031118 | 0.08 |
ENST00000600990.1
|
ZBTB45
|
zinc finger and BTB domain containing 45 |
| chr6_-_133055815 | 0.08 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr1_+_44870866 | 0.08 |
ENST00000355387.2
ENST00000361799.2 |
RNF220
|
ring finger protein 220 |
| chr7_+_139528952 | 0.08 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr4_-_69111401 | 0.08 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr14_-_20922960 | 0.08 |
ENST00000553640.1
ENST00000488532.2 |
OSGEP
|
O-sialoglycoprotein endopeptidase |
| chr4_+_102734967 | 0.08 |
ENST00000444316.2
|
BANK1
|
B-cell scaffold protein with ankyrin repeats 1 |
| chr16_-_3068171 | 0.07 |
ENST00000572154.1
ENST00000328796.4 |
CLDN6
|
claudin 6 |
| chr14_+_91580732 | 0.07 |
ENST00000519019.1
ENST00000523816.1 ENST00000517518.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr17_-_14683517 | 0.07 |
ENST00000379640.1
|
AC005863.1
|
AC005863.1 |
| chr9_+_112852477 | 0.07 |
ENST00000480388.1
|
AKAP2
|
A kinase (PRKA) anchor protein 2 |
| chr8_+_9911945 | 0.07 |
ENST00000518255.1
|
MSRA
|
methionine sulfoxide reductase A |
| chr19_-_55690758 | 0.07 |
ENST00000590851.1
|
SYT5
|
synaptotagmin V |
| chr1_+_28844778 | 0.07 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr15_-_74495188 | 0.07 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr14_+_91580777 | 0.07 |
ENST00000525393.2
ENST00000428926.2 ENST00000517362.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr7_-_148581251 | 0.07 |
ENST00000478654.1
ENST00000460911.1 ENST00000350995.2 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr14_-_73493784 | 0.06 |
ENST00000553891.1
|
ZFYVE1
|
zinc finger, FYVE domain containing 1 |
| chr1_+_244998918 | 0.06 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr4_+_106629929 | 0.06 |
ENST00000512828.1
ENST00000394730.3 ENST00000507281.1 ENST00000515279.1 |
GSTCD
|
glutathione S-transferase, C-terminal domain containing |
| chr9_-_117150303 | 0.06 |
ENST00000312033.3
|
AKNA
|
AT-hook transcription factor |
| chr11_+_46402744 | 0.06 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr9_-_117150243 | 0.06 |
ENST00000374088.3
|
AKNA
|
AT-hook transcription factor |
| chr5_+_93954039 | 0.06 |
ENST00000265140.5
|
ANKRD32
|
ankyrin repeat domain 32 |
| chr11_-_67981295 | 0.06 |
ENST00000405515.1
|
SUV420H1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr10_+_24755416 | 0.06 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr14_+_91580708 | 0.06 |
ENST00000518868.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr2_-_1748214 | 0.06 |
ENST00000433670.1
ENST00000425171.1 ENST00000252804.4 |
PXDN
|
peroxidasin homolog (Drosophila) |
| chr3_-_38992052 | 0.06 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr20_-_52612705 | 0.06 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr5_-_175964366 | 0.06 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chrX_-_128977364 | 0.06 |
ENST00000371064.3
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr15_+_78730622 | 0.06 |
ENST00000560440.1
|
IREB2
|
iron-responsive element binding protein 2 |
| chr1_+_109419804 | 0.06 |
ENST00000435475.1
|
GPSM2
|
G-protein signaling modulator 2 |
| chr5_-_146833222 | 0.06 |
ENST00000534907.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr16_+_14980632 | 0.06 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr19_+_58095501 | 0.06 |
ENST00000536878.2
ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1
|
zinc finger protein interacting with K protein 1 |
| chr2_-_70995307 | 0.06 |
ENST00000264436.4
ENST00000355733.3 ENST00000447731.2 ENST00000430656.1 ENST00000413157.2 |
ADD2
|
adducin 2 (beta) |
| chr7_-_148581360 | 0.06 |
ENST00000320356.2
ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr7_+_30185406 | 0.06 |
ENST00000324489.5
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr16_+_524850 | 0.06 |
ENST00000450428.1
ENST00000452814.1 |
RAB11FIP3
|
RAB11 family interacting protein 3 (class II) |
| chr6_-_139613269 | 0.06 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr6_-_127840048 | 0.05 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr3_+_181429704 | 0.05 |
ENST00000431565.2
ENST00000325404.1 |
SOX2
|
SRY (sex determining region Y)-box 2 |
| chr14_+_69658480 | 0.05 |
ENST00000409949.1
ENST00000409242.1 ENST00000312994.5 ENST00000413191.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chr3_+_111260856 | 0.05 |
ENST00000352690.4
|
CD96
|
CD96 molecule |
| chr19_+_49128209 | 0.05 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
| chr18_-_74728998 | 0.05 |
ENST00000359645.3
ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP
|
myelin basic protein |
| chr11_-_71810258 | 0.05 |
ENST00000544594.1
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr3_+_111260954 | 0.05 |
ENST00000283285.5
|
CD96
|
CD96 molecule |
| chr2_-_227050079 | 0.05 |
ENST00000423838.1
|
AC068138.1
|
AC068138.1 |
| chr7_-_107643567 | 0.05 |
ENST00000393559.1
ENST00000439976.1 ENST00000393560.1 |
LAMB1
|
laminin, beta 1 |
| chr9_+_125795788 | 0.05 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr5_-_146833803 | 0.05 |
ENST00000512722.1
|
DPYSL3
|
dihydropyrimidinase-like 3 |
| chr8_-_95908902 | 0.05 |
ENST00000520509.1
|
CCNE2
|
cyclin E2 |
| chr2_+_11679963 | 0.05 |
ENST00000263834.5
|
GREB1
|
growth regulation by estrogen in breast cancer 1 |
| chr13_+_111893533 | 0.05 |
ENST00000478679.1
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr17_+_9066252 | 0.05 |
ENST00000436734.1
|
NTN1
|
netrin 1 |
| chr14_+_91581011 | 0.05 |
ENST00000523894.1
ENST00000522322.1 ENST00000523771.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr13_+_111855414 | 0.05 |
ENST00000375737.5
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr10_+_51187938 | 0.05 |
ENST00000311663.5
|
FAM21D
|
family with sequence similarity 21, member D |
| chr12_-_62997214 | 0.05 |
ENST00000408887.2
|
C12orf61
|
chromosome 12 open reading frame 61 |
| chr5_+_102200948 | 0.05 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr15_-_34502197 | 0.05 |
ENST00000557877.1
|
KATNBL1
|
katanin p80 subunit B-like 1 |
| chr1_+_153940713 | 0.05 |
ENST00000368601.1
ENST00000368603.1 ENST00000368600.3 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chrX_-_110655391 | 0.04 |
ENST00000356915.2
ENST00000356220.3 |
DCX
|
doublecortin |
| chr15_+_75940218 | 0.04 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr5_+_140739537 | 0.04 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr12_+_132379160 | 0.04 |
ENST00000321867.4
|
ULK1
|
unc-51 like autophagy activating kinase 1 |
| chr4_-_22444733 | 0.04 |
ENST00000508133.1
|
GPR125
|
G protein-coupled receptor 125 |
| chr2_+_201981663 | 0.04 |
ENST00000433445.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_+_93645314 | 0.04 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr15_+_81293254 | 0.04 |
ENST00000267984.2
|
MESDC1
|
mesoderm development candidate 1 |
| chr1_-_153940097 | 0.04 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr12_+_32260085 | 0.04 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr12_+_109915179 | 0.04 |
ENST00000434735.2
|
UBE3B
|
ubiquitin protein ligase E3B |
| chr9_+_139221880 | 0.04 |
ENST00000392945.3
ENST00000440944.1 |
GPSM1
|
G-protein signaling modulator 1 |
| chr8_-_116504448 | 0.04 |
ENST00000518018.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr1_+_209602771 | 0.04 |
ENST00000440276.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr4_+_118955500 | 0.04 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr12_-_99288986 | 0.04 |
ENST00000552407.1
ENST00000551613.1 ENST00000548447.1 ENST00000546364.3 ENST00000552748.1 |
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_+_22146851 | 0.04 |
ENST00000606197.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr12_-_49582837 | 0.04 |
ENST00000547939.1
ENST00000546918.1 ENST00000552924.1 |
TUBA1A
|
tubulin, alpha 1a |
| chr13_+_111855399 | 0.04 |
ENST00000426768.2
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr8_-_6420777 | 0.04 |
ENST00000415216.1
|
ANGPT2
|
angiopoietin 2 |
| chr12_-_49582593 | 0.04 |
ENST00000295766.5
|
TUBA1A
|
tubulin, alpha 1a |
| chr9_+_124088860 | 0.04 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr12_-_95945246 | 0.04 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chrX_+_12993202 | 0.04 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr20_-_56284816 | 0.04 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr5_-_76788317 | 0.04 |
ENST00000296679.4
|
WDR41
|
WD repeat domain 41 |
| chrX_-_24690771 | 0.04 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr14_-_57960456 | 0.04 |
ENST00000534126.1
ENST00000422976.2 |
C14orf105
|
chromosome 14 open reading frame 105 |
| chr20_-_35492048 | 0.04 |
ENST00000237536.4
|
SOGA1
|
suppressor of glucose, autophagy associated 1 |
| chr11_-_62414070 | 0.03 |
ENST00000540933.1
ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB
|
glucosidase, alpha; neutral AB |
| chr16_+_54964740 | 0.03 |
ENST00000394636.4
|
IRX5
|
iroquois homeobox 5 |
| chr10_+_6244829 | 0.03 |
ENST00000317350.4
ENST00000379785.1 ENST00000379782.3 ENST00000360521.2 ENST00000379775.4 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr15_-_72564906 | 0.03 |
ENST00000566844.1
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr6_-_132834184 | 0.03 |
ENST00000367941.2
ENST00000367937.4 |
STX7
|
syntaxin 7 |
| chr22_+_31477296 | 0.03 |
ENST00000426927.1
ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN
|
smoothelin |
| chr4_+_159690218 | 0.03 |
ENST00000264433.6
|
FNIP2
|
folliculin interacting protein 2 |
| chr5_+_140705777 | 0.03 |
ENST00000606901.1
ENST00000606674.1 |
AC005618.6
|
AC005618.6 |
| chr8_+_79578282 | 0.03 |
ENST00000263849.4
|
ZC2HC1A
|
zinc finger, C2HC-type containing 1A |
| chr18_+_3449821 | 0.03 |
ENST00000407501.2
ENST00000405385.3 ENST00000546979.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr6_-_127840021 | 0.03 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr18_+_3450161 | 0.03 |
ENST00000551402.1
ENST00000577543.1 |
TGIF1
|
TGFB-induced factor homeobox 1 |
| chrX_-_73512358 | 0.03 |
ENST00000602776.1
|
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr20_+_30598231 | 0.03 |
ENST00000300415.8
ENST00000262659.8 |
CCM2L
|
cerebral cavernous malformation 2-like |
| chr8_-_6420930 | 0.03 |
ENST00000325203.5
|
ANGPT2
|
angiopoietin 2 |
| chr10_+_73724123 | 0.03 |
ENST00000373115.4
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr12_-_109915098 | 0.03 |
ENST00000542858.1
ENST00000542262.1 ENST00000424763.2 |
KCTD10
|
potassium channel tetramerization domain containing 10 |
| chr4_+_110769258 | 0.03 |
ENST00000594814.1
|
LRIT3
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 |
| chr7_+_141408153 | 0.03 |
ENST00000397541.2
|
WEE2
|
WEE1 homolog 2 (S. pombe) |
| chr1_-_1243392 | 0.03 |
ENST00000354700.5
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr1_+_28261621 | 0.03 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr15_+_25200074 | 0.03 |
ENST00000390687.4
ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN
|
small nuclear ribonucleoprotein polypeptide N |
| chrX_-_110655306 | 0.03 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr17_+_27920486 | 0.03 |
ENST00000394859.3
|
ANKRD13B
|
ankyrin repeat domain 13B |
| chr2_-_20425158 | 0.03 |
ENST00000381150.1
|
SDC1
|
syndecan 1 |
| chrX_+_9880412 | 0.03 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr2_-_114647327 | 0.02 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr22_+_37959647 | 0.02 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr9_-_74979420 | 0.02 |
ENST00000343431.2
ENST00000376956.3 |
ZFAND5
|
zinc finger, AN1-type domain 5 |
| chr3_-_50649192 | 0.02 |
ENST00000443053.2
ENST00000348721.3 |
CISH
|
cytokine inducible SH2-containing protein |
| chr8_-_8751068 | 0.02 |
ENST00000276282.6
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr1_+_153940346 | 0.02 |
ENST00000405694.3
ENST00000449724.1 ENST00000368607.3 ENST00000271889.4 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr22_-_36357671 | 0.02 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr6_+_139456226 | 0.02 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr5_+_102201509 | 0.02 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr7_+_134464376 | 0.02 |
ENST00000454108.1
ENST00000361675.2 |
CALD1
|
caldesmon 1 |
| chr16_-_48281305 | 0.02 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr4_-_87028478 | 0.02 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr6_-_64029879 | 0.02 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chrX_-_153599578 | 0.02 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr1_+_151682909 | 0.02 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr14_+_69658194 | 0.02 |
ENST00000409018.3
ENST00000409014.1 ENST00000409675.1 |
EXD2
|
exonuclease 3'-5' domain containing 2 |
| chrX_-_69509738 | 0.02 |
ENST00000374454.1
ENST00000239666.4 |
PDZD11
|
PDZ domain containing 11 |
| chr17_+_64961026 | 0.02 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr7_+_134464414 | 0.02 |
ENST00000361901.2
|
CALD1
|
caldesmon 1 |
| chr11_-_10830463 | 0.02 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr12_+_69004619 | 0.02 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.3 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.0 | GO:1900737 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.0 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.0 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0043219 | lateral loop(GO:0043219) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.2 | GO:0072591 | citrate-L-glutamate ligase activity(GO:0072591) |
| 0.0 | 0.3 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.3 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |