Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
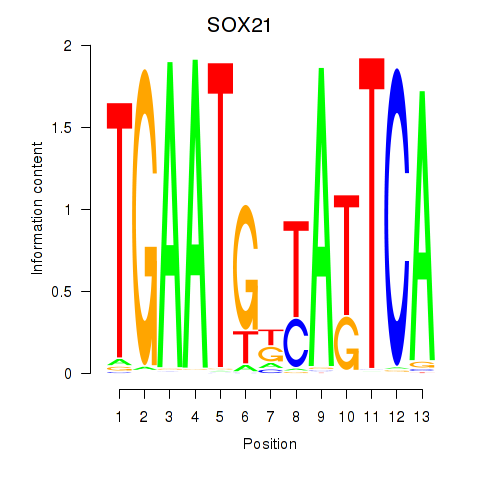
Results for SOX21
Z-value: 0.70
Transcription factors associated with SOX21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX21
|
ENSG00000125285.4 | SRY-box transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX21 | hg19_v2_chr13_-_95364389_95364389 | -0.03 | 9.6e-01 | Click! |
Activity profile of SOX21 motif
Sorted Z-values of SOX21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_101361107 | 0.66 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chr19_+_42381337 | 0.47 |
ENST00000597454.1
ENST00000444740.2 |
CD79A
|
CD79a molecule, immunoglobulin-associated alpha |
| chr10_-_18944123 | 0.36 |
ENST00000606425.1
|
RP11-139J15.7
|
Uncharacterized protein |
| chr3_-_37034702 | 0.31 |
ENST00000322716.5
|
EPM2AIP1
|
EPM2A (laforin) interacting protein 1 |
| chr9_-_70465758 | 0.29 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr4_-_39034542 | 0.28 |
ENST00000344606.6
|
TMEM156
|
transmembrane protein 156 |
| chr14_+_64565442 | 0.26 |
ENST00000553308.1
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chr12_-_10588539 | 0.24 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chr3_+_38537960 | 0.24 |
ENST00000453767.1
|
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr13_-_45048386 | 0.21 |
ENST00000472477.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr3_-_45957088 | 0.21 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr8_-_94029882 | 0.21 |
ENST00000520686.1
|
TRIQK
|
triple QxxK/R motif containing |
| chrY_+_14813160 | 0.20 |
ENST00000338981.3
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr5_+_67588312 | 0.18 |
ENST00000519025.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr7_-_86849883 | 0.18 |
ENST00000433078.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr8_+_74271144 | 0.17 |
ENST00000519134.1
ENST00000518355.1 |
RP11-434I12.3
|
RP11-434I12.3 |
| chr7_+_7196565 | 0.16 |
ENST00000429911.1
|
C1GALT1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr3_-_45957534 | 0.16 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr3_-_155524049 | 0.15 |
ENST00000534941.1
ENST00000340171.2 |
C3orf33
|
chromosome 3 open reading frame 33 |
| chr18_+_29769978 | 0.15 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr14_+_20215587 | 0.15 |
ENST00000331723.1
|
OR4Q3
|
olfactory receptor, family 4, subfamily Q, member 3 |
| chr6_-_121655593 | 0.15 |
ENST00000398212.2
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr4_-_103747011 | 0.14 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_-_17243579 | 0.14 |
ENST00000525762.1
ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1
|
tRNA aspartic acid methyltransferase 1 |
| chr5_-_95158644 | 0.14 |
ENST00000237858.6
|
GLRX
|
glutaredoxin (thioltransferase) |
| chr10_-_71169031 | 0.14 |
ENST00000373307.1
|
TACR2
|
tachykinin receptor 2 |
| chr1_+_158901329 | 0.13 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr14_+_85994943 | 0.13 |
ENST00000553678.1
|
RP11-497E19.2
|
Uncharacterized protein |
| chr1_-_169455169 | 0.13 |
ENST00000367804.4
ENST00000236137.5 |
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr15_+_58702742 | 0.12 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr4_-_103746924 | 0.12 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_38537763 | 0.12 |
ENST00000287675.5
ENST00000358249.2 ENST00000422077.2 |
EXOG
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr1_-_54303934 | 0.12 |
ENST00000537333.1
|
NDC1
|
NDC1 transmembrane nucleoporin |
| chr12_+_123011776 | 0.12 |
ENST00000450485.2
ENST00000333479.7 |
KNTC1
|
kinetochore associated 1 |
| chr2_-_106013207 | 0.11 |
ENST00000447958.1
|
FHL2
|
four and a half LIM domains 2 |
| chr4_-_103746683 | 0.11 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr10_-_12084770 | 0.11 |
ENST00000357604.5
|
UPF2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr3_-_164875850 | 0.11 |
ENST00000472120.1
|
RP11-747D18.1
|
RP11-747D18.1 |
| chr1_+_199996733 | 0.11 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr11_+_114549108 | 0.10 |
ENST00000389586.4
ENST00000375475.5 |
NXPE2
|
neurexophilin and PC-esterase domain family, member 2 |
| chr10_+_5005445 | 0.10 |
ENST00000380872.4
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr6_+_42531798 | 0.10 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr17_+_66245341 | 0.09 |
ENST00000577985.1
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr6_+_131894284 | 0.08 |
ENST00000368087.3
ENST00000356962.2 |
ARG1
|
arginase 1 |
| chr3_-_155572164 | 0.08 |
ENST00000392845.3
ENST00000359479.3 |
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr7_+_6713376 | 0.08 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr11_-_8285405 | 0.08 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr1_+_221051699 | 0.07 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr5_-_95158375 | 0.07 |
ENST00000512469.2
ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX
|
glutaredoxin (thioltransferase) |
| chr16_-_15149828 | 0.07 |
ENST00000566419.1
ENST00000568320.1 |
NTAN1
|
N-terminal asparagine amidase |
| chr1_-_151148492 | 0.07 |
ENST00000295314.4
|
TMOD4
|
tropomodulin 4 (muscle) |
| chr10_-_61495760 | 0.07 |
ENST00000395347.1
|
SLC16A9
|
solute carrier family 16, member 9 |
| chr7_-_37382683 | 0.06 |
ENST00000455879.1
|
ELMO1
|
engulfment and cell motility 1 |
| chrX_-_153210107 | 0.06 |
ENST00000369997.3
ENST00000393700.3 ENST00000412763.1 |
RENBP
|
renin binding protein |
| chr1_+_152881014 | 0.06 |
ENST00000368764.3
ENST00000392667.2 |
IVL
|
involucrin |
| chr14_-_92588013 | 0.06 |
ENST00000553514.1
ENST00000605997.1 |
NDUFB1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr9_+_109685630 | 0.06 |
ENST00000451160.2
|
RP11-508N12.4
|
Uncharacterized protein |
| chr5_+_140579162 | 0.06 |
ENST00000536699.1
ENST00000354757.3 |
PCDHB11
|
protocadherin beta 11 |
| chr18_-_47018897 | 0.05 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr16_-_15982440 | 0.05 |
ENST00000575938.1
ENST00000573396.1 ENST00000573968.1 ENST00000575744.1 ENST00000573429.1 ENST00000255759.6 ENST00000575073.1 |
FOPNL
|
FGFR1OP N-terminal like |
| chr12_+_101673872 | 0.05 |
ENST00000261637.4
|
UTP20
|
UTP20, small subunit (SSU) processome component, homolog (yeast) |
| chr8_-_50466973 | 0.04 |
ENST00000520800.1
|
RP11-738G5.2
|
Uncharacterized protein |
| chr6_-_154567984 | 0.04 |
ENST00000517438.1
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1 |
| chr8_+_26150628 | 0.04 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr17_+_60457914 | 0.04 |
ENST00000305286.3
ENST00000520404.1 ENST00000518576.1 |
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr22_+_39353527 | 0.04 |
ENST00000249116.2
|
APOBEC3A
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A |
| chr14_+_92588281 | 0.04 |
ENST00000298875.4
ENST00000553427.1 |
CPSF2
|
cleavage and polyadenylation specific factor 2, 100kDa |
| chr14_-_92588246 | 0.04 |
ENST00000329559.3
|
NDUFB1
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa |
| chr12_+_69742121 | 0.04 |
ENST00000261267.2
ENST00000549690.1 ENST00000548839.1 |
LYZ
|
lysozyme |
| chr14_+_50291993 | 0.03 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr3_-_141747439 | 0.03 |
ENST00000467667.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chrY_+_16634483 | 0.03 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr5_+_134181755 | 0.03 |
ENST00000504727.1
ENST00000435259.2 ENST00000508791.1 |
C5orf24
|
chromosome 5 open reading frame 24 |
| chr18_-_47018769 | 0.03 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr6_+_31371337 | 0.02 |
ENST00000449934.2
ENST00000421350.1 |
MICA
|
MHC class I polypeptide-related sequence A |
| chr12_+_59194154 | 0.02 |
ENST00000548969.1
|
RP11-362K2.2
|
Protein LOC100506869 |
| chr20_+_56964253 | 0.02 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr18_+_43319467 | 0.02 |
ENST00000591541.1
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr6_-_112575912 | 0.02 |
ENST00000522006.1
ENST00000230538.7 ENST00000519932.1 |
LAMA4
|
laminin, alpha 4 |
| chr3_-_186288097 | 0.02 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr2_+_3705785 | 0.01 |
ENST00000252505.3
|
ALLC
|
allantoicase |
| chr18_-_47018869 | 0.01 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr1_+_203764742 | 0.01 |
ENST00000432282.1
ENST00000453771.1 ENST00000367214.1 ENST00000367212.3 ENST00000332127.4 |
ZC3H11A
|
zinc finger CCCH-type containing 11A |
| chr19_+_18682531 | 0.01 |
ENST00000596304.1
ENST00000430157.2 |
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr1_+_67632083 | 0.01 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr5_-_179045199 | 0.01 |
ENST00000523921.1
|
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr9_-_104249400 | 0.00 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246 |
| chr8_+_67687413 | 0.00 |
ENST00000521960.1
ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr7_-_107968921 | 0.00 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr21_+_30672433 | 0.00 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr8_-_66750978 | 0.00 |
ENST00000523253.1
|
PDE7A
|
phosphodiesterase 7A |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX21
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903568 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.2 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:0030474 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.3 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.0 | 0.0 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.1 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |