Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
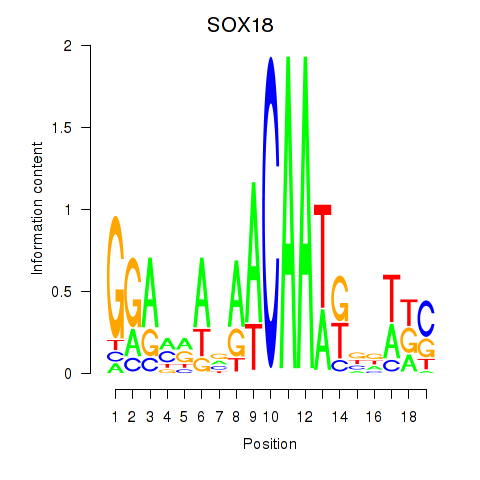
Results for SOX18
Z-value: 0.70
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.5 | SRY-box transcription factor 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg19_v2_chr20_-_62680984_62680999 | 0.36 | 4.9e-01 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26235206 | 0.86 |
ENST00000244534.5
|
HIST1H1D
|
histone cluster 1, H1d |
| chr22_-_38480100 | 0.58 |
ENST00000427592.1
|
SLC16A8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr8_-_1922789 | 0.50 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr12_-_66317967 | 0.41 |
ENST00000601398.1
|
AC090673.2
|
Uncharacterized protein |
| chr12_-_11244912 | 0.41 |
ENST00000531678.1
|
TAS2R43
|
taste receptor, type 2, member 43 |
| chr12_+_11905413 | 0.41 |
ENST00000545027.1
|
ETV6
|
ets variant 6 |
| chr5_+_127039075 | 0.35 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr12_-_85430024 | 0.35 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr11_+_123986069 | 0.30 |
ENST00000456829.2
ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A
|
von Willebrand factor A domain containing 5A |
| chr5_-_99870932 | 0.30 |
ENST00000504833.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr6_-_27880174 | 0.29 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr12_-_11002063 | 0.28 |
ENST00000544994.1
ENST00000228811.4 ENST00000540107.1 |
PRR4
|
proline rich 4 (lacrimal) |
| chr20_-_44718538 | 0.26 |
ENST00000290231.6
ENST00000372291.3 |
NCOA5
|
nuclear receptor coactivator 5 |
| chr11_+_327171 | 0.25 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr19_-_46105411 | 0.25 |
ENST00000323040.4
ENST00000544371.1 |
GPR4
OPA3
|
G protein-coupled receptor 4 optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr22_-_31324215 | 0.23 |
ENST00000429468.1
|
MORC2
|
MORC family CW-type zinc finger 2 |
| chr11_+_5711010 | 0.21 |
ENST00000454828.1
|
TRIM22
|
tripartite motif containing 22 |
| chr8_+_23145594 | 0.20 |
ENST00000519952.1
ENST00000518840.1 |
R3HCC1
|
R3H domain and coiled-coil containing 1 |
| chr2_+_101591314 | 0.20 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr12_+_70574088 | 0.20 |
ENST00000552324.1
|
RP11-320P7.2
|
RP11-320P7.2 |
| chr17_-_28661065 | 0.20 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr8_-_37707356 | 0.19 |
ENST00000520601.1
ENST00000521170.1 ENST00000220659.6 |
BRF2
|
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chr4_-_100484825 | 0.19 |
ENST00000273962.3
ENST00000514547.1 ENST00000455368.2 |
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chrX_+_10031499 | 0.19 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr13_+_110958124 | 0.18 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr3_+_187420101 | 0.17 |
ENST00000449623.1
ENST00000437407.1 |
RP11-211G3.3
|
Uncharacterized protein |
| chr12_-_70093235 | 0.17 |
ENST00000266661.4
|
BEST3
|
bestrophin 3 |
| chr9_-_25678856 | 0.17 |
ENST00000358022.3
|
TUSC1
|
tumor suppressor candidate 1 |
| chr5_-_67730240 | 0.17 |
ENST00000507733.1
|
CTC-537E7.3
|
CTC-537E7.3 |
| chr12_+_85430110 | 0.17 |
ENST00000393212.3
ENST00000393217.2 |
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1 |
| chr11_+_74699942 | 0.16 |
ENST00000526068.1
ENST00000532963.1 ENST00000531619.1 ENST00000534628.1 ENST00000545272.1 |
NEU3
|
sialidase 3 (membrane sialidase) |
| chr11_-_76155618 | 0.16 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr16_-_28550320 | 0.16 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr1_+_15256230 | 0.15 |
ENST00000376028.4
ENST00000400798.2 |
KAZN
|
kazrin, periplakin interacting protein |
| chr13_-_46716969 | 0.15 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chrX_+_47077632 | 0.15 |
ENST00000457458.2
|
CDK16
|
cyclin-dependent kinase 16 |
| chr17_+_48503519 | 0.15 |
ENST00000300441.4
ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr17_-_76719638 | 0.14 |
ENST00000587308.1
|
CYTH1
|
cytohesin 1 |
| chr2_-_105953912 | 0.14 |
ENST00000610036.1
|
RP11-332H14.2
|
RP11-332H14.2 |
| chr2_+_74881432 | 0.14 |
ENST00000453930.1
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr6_-_88001706 | 0.14 |
ENST00000369576.2
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr5_-_146461027 | 0.14 |
ENST00000394410.2
ENST00000508267.1 ENST00000504198.1 |
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta |
| chrX_+_47078069 | 0.13 |
ENST00000357227.4
ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16
|
cyclin-dependent kinase 16 |
| chr2_-_47382414 | 0.12 |
ENST00000294947.2
|
C2orf61
|
chromosome 2 open reading frame 61 |
| chr16_-_54972581 | 0.12 |
ENST00000559802.1
ENST00000558156.1 |
CTD-3032H12.1
|
CTD-3032H12.1 |
| chr15_-_30261066 | 0.12 |
ENST00000558447.1
|
TJP1
|
tight junction protein 1 |
| chr17_+_12569472 | 0.12 |
ENST00000343344.4
|
MYOCD
|
myocardin |
| chr8_-_123793048 | 0.12 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr8_+_97773457 | 0.12 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr19_+_41256764 | 0.12 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr22_+_20850171 | 0.11 |
ENST00000445987.1
|
MED15
|
mediator complex subunit 15 |
| chr3_-_197300194 | 0.11 |
ENST00000358186.2
ENST00000431056.1 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr17_+_48503603 | 0.11 |
ENST00000502667.1
|
ACSF2
|
acyl-CoA synthetase family member 2 |
| chr19_-_39805976 | 0.11 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr3_+_33331156 | 0.11 |
ENST00000542085.1
|
FBXL2
|
F-box and leucine-rich repeat protein 2 |
| chr3_-_127309521 | 0.11 |
ENST00000469111.1
|
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chr17_+_38516907 | 0.11 |
ENST00000578774.1
|
CTD-2267D19.3
|
Uncharacterized protein |
| chr6_-_110500826 | 0.11 |
ENST00000265601.3
ENST00000447287.1 ENST00000444391.1 |
WASF1
|
WAS protein family, member 1 |
| chr19_+_4402659 | 0.11 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr8_+_24151620 | 0.11 |
ENST00000437154.2
|
ADAM28
|
ADAM metallopeptidase domain 28 |
| chr10_-_127505167 | 0.11 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr11_+_113258495 | 0.11 |
ENST00000303941.3
|
ANKK1
|
ankyrin repeat and kinase domain containing 1 |
| chr9_-_128246769 | 0.11 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr6_-_33754778 | 0.11 |
ENST00000508327.1
ENST00000513701.1 |
LEMD2
|
LEM domain containing 2 |
| chr12_-_67197760 | 0.11 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr8_-_13134045 | 0.11 |
ENST00000512044.2
|
DLC1
|
deleted in liver cancer 1 |
| chrX_-_154033661 | 0.11 |
ENST00000393531.1
|
MPP1
|
membrane protein, palmitoylated 1, 55kDa |
| chr15_-_59500973 | 0.10 |
ENST00000560749.1
|
MYO1E
|
myosin IE |
| chr7_-_45026419 | 0.10 |
ENST00000578968.1
ENST00000580528.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr7_-_117512264 | 0.10 |
ENST00000454375.1
|
CTTNBP2
|
cortactin binding protein 2 |
| chrX_+_119495934 | 0.10 |
ENST00000218008.3
ENST00000361319.3 ENST00000539306.1 |
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr5_-_39462390 | 0.10 |
ENST00000511792.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr22_+_37678424 | 0.10 |
ENST00000248901.6
|
CYTH4
|
cytohesin 4 |
| chr10_-_73497581 | 0.10 |
ENST00000398786.2
|
C10orf105
|
chromosome 10 open reading frame 105 |
| chr2_-_158345341 | 0.10 |
ENST00000435117.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr2_+_74881398 | 0.10 |
ENST00000339773.5
ENST00000434486.1 |
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr15_-_41166414 | 0.10 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr1_-_114430169 | 0.09 |
ENST00000393316.3
|
BCL2L15
|
BCL2-like 15 |
| chr11_-_118966167 | 0.09 |
ENST00000530167.1
|
H2AFX
|
H2A histone family, member X |
| chr15_+_82380974 | 0.09 |
ENST00000559788.1
ENST00000557838.1 |
RP11-597K23.2
|
Uncharacterized protein |
| chr6_-_31125850 | 0.09 |
ENST00000507751.1
ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1
|
coiled-coil alpha-helical rod protein 1 |
| chr12_-_56321397 | 0.09 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr2_+_74881355 | 0.09 |
ENST00000357877.2
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr12_+_110338323 | 0.09 |
ENST00000312777.5
ENST00000536408.2 |
TCHP
|
trichoplein, keratin filament binding |
| chr16_-_787771 | 0.09 |
ENST00000568545.1
|
NARFL
|
nuclear prelamin A recognition factor-like |
| chr6_+_26217159 | 0.09 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr15_+_67418047 | 0.09 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr10_-_13390270 | 0.09 |
ENST00000378614.4
ENST00000545675.1 ENST00000327347.5 |
SEPHS1
|
selenophosphate synthetase 1 |
| chr4_-_114900831 | 0.09 |
ENST00000315366.7
|
ARSJ
|
arylsulfatase family, member J |
| chr6_-_152489484 | 0.09 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr11_+_64949343 | 0.09 |
ENST00000279247.6
ENST00000532285.1 ENST00000534373.1 |
CAPN1
|
calpain 1, (mu/I) large subunit |
| chr5_-_31532160 | 0.09 |
ENST00000511367.2
ENST00000513349.1 |
DROSHA
|
drosha, ribonuclease type III |
| chr16_+_28770025 | 0.08 |
ENST00000435324.2
|
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr2_-_220197351 | 0.08 |
ENST00000392083.1
|
RESP18
|
regulated endocrine-specific protein 18 |
| chr11_+_73358690 | 0.08 |
ENST00000545798.1
ENST00000539157.1 ENST00000546251.1 ENST00000535582.1 ENST00000538227.1 ENST00000543524.1 |
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chrX_-_49041242 | 0.08 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr19_+_33571786 | 0.08 |
ENST00000170564.2
|
GPATCH1
|
G patch domain containing 1 |
| chr2_+_1418154 | 0.08 |
ENST00000423320.1
ENST00000382198.1 |
TPO
|
thyroid peroxidase |
| chr7_-_99716940 | 0.08 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr4_+_54243798 | 0.08 |
ENST00000337488.6
ENST00000358575.5 ENST00000507922.1 |
FIP1L1
|
factor interacting with PAPOLA and CPSF1 |
| chr3_-_47932759 | 0.08 |
ENST00000441748.2
ENST00000335271.5 |
MAP4
|
microtubule-associated protein 4 |
| chr1_+_43124087 | 0.08 |
ENST00000304979.3
ENST00000372550.1 ENST00000440068.1 |
PPIH
|
peptidylprolyl isomerase H (cyclophilin H) |
| chr17_+_25799008 | 0.08 |
ENST00000583370.1
ENST00000398988.3 ENST00000268763.6 |
KSR1
|
kinase suppressor of ras 1 |
| chr3_+_57882024 | 0.08 |
ENST00000494088.1
|
SLMAP
|
sarcolemma associated protein |
| chr7_+_99933730 | 0.08 |
ENST00000610247.1
|
PILRB
|
paired immunoglobin-like type 2 receptor beta |
| chr15_+_91473403 | 0.08 |
ENST00000394275.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr11_-_76155700 | 0.08 |
ENST00000572035.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr12_+_32260085 | 0.08 |
ENST00000548411.1
ENST00000281474.5 ENST00000551086.1 |
BICD1
|
bicaudal D homolog 1 (Drosophila) |
| chr6_-_33385655 | 0.08 |
ENST00000440279.3
ENST00000607266.1 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr1_-_201123546 | 0.08 |
ENST00000435310.1
ENST00000485839.2 ENST00000367330.1 |
TMEM9
|
transmembrane protein 9 |
| chr16_-_4896205 | 0.08 |
ENST00000589389.1
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr15_+_75628232 | 0.07 |
ENST00000267935.8
ENST00000567195.1 |
COMMD4
|
COMM domain containing 4 |
| chrX_+_47004639 | 0.07 |
ENST00000345781.6
|
RBM10
|
RNA binding motif protein 10 |
| chr7_-_148725733 | 0.07 |
ENST00000286091.4
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr11_-_6677018 | 0.07 |
ENST00000299441.3
|
DCHS1
|
dachsous cadherin-related 1 |
| chr15_+_69857515 | 0.07 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chrX_+_100645812 | 0.07 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr12_-_11139511 | 0.07 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr6_-_33385870 | 0.07 |
ENST00000488034.1
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr7_-_122840015 | 0.07 |
ENST00000194130.2
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr16_-_68034470 | 0.07 |
ENST00000412757.2
|
DPEP2
|
dipeptidase 2 |
| chr2_-_47382442 | 0.07 |
ENST00000445927.2
|
C2orf61
|
chromosome 2 open reading frame 61 |
| chr6_-_33385823 | 0.06 |
ENST00000494751.1
ENST00000374496.3 |
CUTA
|
cutA divalent cation tolerance homolog (E. coli) |
| chr9_+_137987825 | 0.06 |
ENST00000545657.1
|
OLFM1
|
olfactomedin 1 |
| chr14_+_24458123 | 0.06 |
ENST00000545240.1
ENST00000382755.4 |
DHRS4L2
|
dehydrogenase/reductase (SDR family) member 4 like 2 |
| chr3_+_145782358 | 0.06 |
ENST00000422482.1
|
AC107021.1
|
HCG1786590; PRO2533; Uncharacterized protein |
| chr12_-_371994 | 0.06 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr2_-_165811756 | 0.06 |
ENST00000409662.1
|
SLC38A11
|
solute carrier family 38, member 11 |
| chr1_+_36348790 | 0.06 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr6_+_25652501 | 0.06 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr2_-_208031943 | 0.06 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr5_+_57878859 | 0.06 |
ENST00000282878.4
|
RAB3C
|
RAB3C, member RAS oncogene family |
| chr5_-_55412774 | 0.06 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chrX_-_47004437 | 0.06 |
ENST00000276062.8
|
NDUFB11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr19_+_41257084 | 0.06 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chrX_+_47004599 | 0.06 |
ENST00000329236.7
|
RBM10
|
RNA binding motif protein 10 |
| chr15_+_64386261 | 0.06 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chrX_+_36254051 | 0.06 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr9_+_125437315 | 0.06 |
ENST00000304820.2
|
OR1L3
|
olfactory receptor, family 1, subfamily L, member 3 |
| chr12_+_110338063 | 0.06 |
ENST00000405876.4
|
TCHP
|
trichoplein, keratin filament binding |
| chr3_+_190281229 | 0.06 |
ENST00000453359.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr3_+_57881966 | 0.06 |
ENST00000495364.1
|
SLMAP
|
sarcolemma associated protein |
| chr1_+_73771844 | 0.06 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr19_+_39971505 | 0.06 |
ENST00000544017.1
|
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr16_-_79804394 | 0.06 |
ENST00000567993.1
|
RP11-345M22.2
|
RP11-345M22.2 |
| chr12_-_27235447 | 0.05 |
ENST00000429849.2
|
C12orf71
|
chromosome 12 open reading frame 71 |
| chr3_+_35721182 | 0.05 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_-_206785789 | 0.05 |
ENST00000437518.1
ENST00000367114.3 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr17_+_41158742 | 0.05 |
ENST00000415816.2
ENST00000438323.2 |
IFI35
|
interferon-induced protein 35 |
| chr1_-_157746909 | 0.05 |
ENST00000392274.3
ENST00000361516.3 ENST00000368181.4 |
FCRL2
|
Fc receptor-like 2 |
| chr19_+_35842445 | 0.05 |
ENST00000246553.2
|
FFAR1
|
free fatty acid receptor 1 |
| chr12_+_96252706 | 0.05 |
ENST00000266735.5
ENST00000553192.1 ENST00000552085.1 |
SNRPF
|
small nuclear ribonucleoprotein polypeptide F |
| chr7_-_45026200 | 0.05 |
ENST00000577700.1
ENST00000580458.1 ENST00000579383.1 ENST00000584686.1 ENST00000585030.1 ENST00000582727.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr17_+_41150290 | 0.05 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr2_+_210518057 | 0.05 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr19_-_7936344 | 0.05 |
ENST00000599142.1
|
CTD-3193O13.9
|
Protein FLJ22184 |
| chr6_+_123317116 | 0.05 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr2_+_217277137 | 0.05 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr17_+_41150479 | 0.05 |
ENST00000589913.1
|
RPL27
|
ribosomal protein L27 |
| chrX_+_52920336 | 0.05 |
ENST00000452154.2
|
FAM156B
|
family with sequence similarity 156, member B |
| chr18_+_3447572 | 0.05 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr3_+_57741957 | 0.05 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr11_+_13299186 | 0.05 |
ENST00000527998.1
ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL
|
aryl hydrocarbon receptor nuclear translocator-like |
| chr13_-_46756351 | 0.05 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_+_10459433 | 0.05 |
ENST00000465632.1
ENST00000460189.1 |
PGD
|
phosphogluconate dehydrogenase |
| chr22_+_31488433 | 0.05 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr8_+_11225911 | 0.05 |
ENST00000284481.3
|
C8orf12
|
chromosome 8 open reading frame 12 |
| chr1_+_161123536 | 0.05 |
ENST00000368003.5
|
UFC1
|
ubiquitin-fold modifier conjugating enzyme 1 |
| chr8_+_35649365 | 0.05 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr19_+_39971470 | 0.05 |
ENST00000607714.1
ENST00000599794.1 ENST00000597666.1 ENST00000601403.1 ENST00000602028.1 |
TIMM50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr12_+_69633407 | 0.05 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr5_-_99870890 | 0.05 |
ENST00000499025.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr5_+_32711829 | 0.05 |
ENST00000415167.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr1_+_33231221 | 0.05 |
ENST00000294521.3
|
KIAA1522
|
KIAA1522 |
| chr1_-_206785898 | 0.05 |
ENST00000271764.2
|
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr7_-_6048650 | 0.05 |
ENST00000382321.4
ENST00000406569.3 |
PMS2
|
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr3_-_121379739 | 0.05 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr11_-_34379546 | 0.05 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr19_-_43382142 | 0.05 |
ENST00000597058.1
|
PSG1
|
pregnancy specific beta-1-glycoprotein 1 |
| chr17_-_39123144 | 0.05 |
ENST00000355612.2
|
KRT39
|
keratin 39 |
| chr8_+_97773202 | 0.05 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr3_-_184971853 | 0.04 |
ENST00000231887.3
|
EHHADH
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr15_+_44084040 | 0.04 |
ENST00000249786.4
|
SERF2
|
small EDRK-rich factor 2 |
| chr19_+_44576296 | 0.04 |
ENST00000421176.3
|
ZNF284
|
zinc finger protein 284 |
| chr12_-_48597170 | 0.04 |
ENST00000310248.2
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1 |
| chr1_-_201123586 | 0.04 |
ENST00000414605.2
ENST00000367334.5 ENST00000367332.1 |
TMEM9
|
transmembrane protein 9 |
| chr1_-_153513170 | 0.04 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr6_-_31689456 | 0.04 |
ENST00000495859.1
ENST00000375819.2 |
LY6G6C
|
lymphocyte antigen 6 complex, locus G6C |
| chr10_-_135187193 | 0.04 |
ENST00000368547.3
|
ECHS1
|
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr19_+_1249869 | 0.04 |
ENST00000591446.2
|
MIDN
|
midnolin |
| chr12_+_3000073 | 0.04 |
ENST00000397132.2
|
TULP3
|
tubby like protein 3 |
| chr12_+_113344755 | 0.04 |
ENST00000550883.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr3_+_187896331 | 0.04 |
ENST00000392468.2
|
AC022498.1
|
Uncharacterized protein |
| chr13_-_24471641 | 0.04 |
ENST00000382145.1
|
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr13_-_33780133 | 0.04 |
ENST00000399365.3
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13 |
| chr15_+_28624878 | 0.04 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr11_+_5710919 | 0.04 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr1_-_32801825 | 0.04 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chrX_+_47053208 | 0.04 |
ENST00000442035.1
ENST00000457753.1 ENST00000335972.6 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr12_-_11150474 | 0.03 |
ENST00000538986.1
|
TAS2R20
|
taste receptor, type 2, member 20 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0044727 | DNA demethylation of male pronucleus(GO:0044727) |
| 0.0 | 0.6 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:1900276 | regulation of proteinase activated receptor activity(GO:1900276) negative regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900737) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.6 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.0 | GO:0019520 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:1901671 | positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.0 | 0.0 | GO:2000627 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0039713 | viral factory(GO:0039713) cytoplasmic viral factory(GO:0039714) host cell viral assembly compartment(GO:0072517) |
| 0.0 | 0.1 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.9 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.2 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |