Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
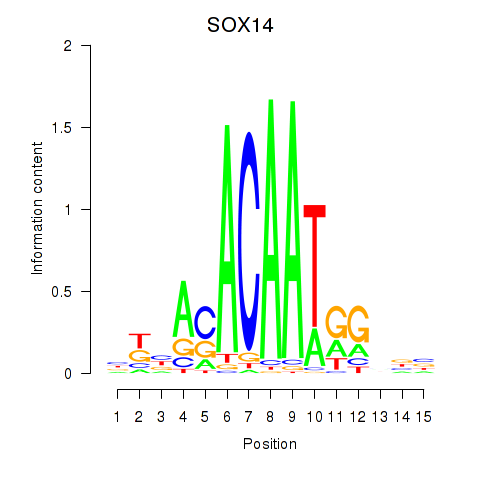
Results for SOX14
Z-value: 0.76
Transcription factors associated with SOX14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX14
|
ENSG00000168875.1 | SRY-box transcription factor 14 |
Activity profile of SOX14 motif
Sorted Z-values of SOX14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_113354341 | 1.11 |
ENST00000553152.1
|
OAS1
|
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr5_+_111755280 | 1.05 |
ENST00000600409.1
|
EPB41L4A-AS2
|
EPB41L4A antisense RNA 2 (head to head) |
| chr14_+_24641062 | 0.67 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr11_+_17316870 | 0.48 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr6_+_26045603 | 0.48 |
ENST00000540144.1
|
HIST1H3C
|
histone cluster 1, H3c |
| chr12_+_65996599 | 0.47 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr15_+_96897466 | 0.44 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chr12_+_69633407 | 0.38 |
ENST00000551516.1
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr3_+_164924716 | 0.37 |
ENST00000470138.1
ENST00000498616.1 |
RP11-85M11.2
|
RP11-85M11.2 |
| chr12_+_95611569 | 0.37 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr15_+_71184931 | 0.36 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr16_+_30934376 | 0.34 |
ENST00000562798.1
ENST00000471231.2 |
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr22_-_50970506 | 0.33 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr6_+_138188551 | 0.33 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr7_-_149470540 | 0.32 |
ENST00000302017.3
|
ZNF467
|
zinc finger protein 467 |
| chr19_-_17375541 | 0.32 |
ENST00000252597.3
|
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chr1_+_14026722 | 0.31 |
ENST00000376048.5
|
PRDM2
|
PR domain containing 2, with ZNF domain |
| chr22_+_24115000 | 0.31 |
ENST00000215743.3
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3) |
| chr17_-_42441204 | 0.31 |
ENST00000293443.7
|
FAM171A2
|
family with sequence similarity 171, member A2 |
| chr17_-_40337470 | 0.29 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr6_+_29691198 | 0.26 |
ENST00000440587.2
ENST00000434407.2 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr11_-_63684316 | 0.26 |
ENST00000301459.4
|
RCOR2
|
REST corepressor 2 |
| chr5_-_111754948 | 0.25 |
ENST00000261486.5
|
EPB41L4A
|
erythrocyte membrane protein band 4.1 like 4A |
| chr20_-_62493217 | 0.25 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr1_+_52521957 | 0.25 |
ENST00000472944.2
ENST00000484036.1 |
BTF3L4
|
basic transcription factor 3-like 4 |
| chr17_-_18585131 | 0.24 |
ENST00000443457.1
ENST00000583002.1 |
ZNF286B
|
zinc finger protein 286B |
| chr6_+_29691056 | 0.24 |
ENST00000414333.1
ENST00000334668.4 ENST00000259951.7 |
HLA-F
|
major histocompatibility complex, class I, F |
| chr12_+_95611536 | 0.24 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr4_-_103266626 | 0.23 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr12_+_95611516 | 0.23 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr12_+_498500 | 0.22 |
ENST00000540180.1
ENST00000422000.1 ENST00000535052.1 |
CCDC77
|
coiled-coil domain containing 77 |
| chr17_-_79106634 | 0.22 |
ENST00000575363.1
|
AATK
|
apoptosis-associated tyrosine kinase |
| chr2_+_170590321 | 0.22 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr4_+_26323764 | 0.21 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr10_+_30722866 | 0.20 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr12_-_49581152 | 0.20 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr1_+_65886326 | 0.19 |
ENST00000371059.3
ENST00000371060.3 ENST00000349533.6 ENST00000406510.3 |
LEPR
|
leptin receptor |
| chrX_-_138914394 | 0.19 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr4_-_164265984 | 0.19 |
ENST00000511901.1
|
NPY1R
|
neuropeptide Y receptor Y1 |
| chr1_-_31712401 | 0.19 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr7_-_105925558 | 0.19 |
ENST00000222553.3
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr2_+_240323439 | 0.18 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr17_+_39411636 | 0.18 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr15_+_52311398 | 0.17 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr3_+_49840685 | 0.17 |
ENST00000333323.4
|
FAM212A
|
family with sequence similarity 212, member A |
| chr14_+_103589789 | 0.17 |
ENST00000558056.1
ENST00000560869.1 |
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2 |
| chr10_-_33247124 | 0.17 |
ENST00000414670.1
ENST00000302278.3 ENST00000374956.4 ENST00000488494.1 ENST00000417122.2 ENST00000474568.1 |
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr6_-_86353510 | 0.17 |
ENST00000444272.1
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr10_+_121578211 | 0.17 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr12_-_52887034 | 0.16 |
ENST00000330722.6
|
KRT6A
|
keratin 6A |
| chr14_-_35344093 | 0.16 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr3_+_48507210 | 0.15 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr2_-_160472052 | 0.15 |
ENST00000437839.1
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr11_-_104905840 | 0.15 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr1_+_40915725 | 0.15 |
ENST00000484445.1
ENST00000411995.2 ENST00000361584.3 |
ZFP69B
|
ZFP69 zinc finger protein B |
| chr2_+_187371440 | 0.14 |
ENST00000445547.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr17_+_4487294 | 0.14 |
ENST00000338859.4
|
SMTNL2
|
smoothelin-like 2 |
| chr2_+_153191706 | 0.14 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr17_+_7608511 | 0.14 |
ENST00000226091.2
|
EFNB3
|
ephrin-B3 |
| chr10_+_70480963 | 0.14 |
ENST00000265872.6
ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1
|
cell division cycle and apoptosis regulator 1 |
| chr4_+_113152978 | 0.13 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr12_+_498545 | 0.13 |
ENST00000543504.1
|
CCDC77
|
coiled-coil domain containing 77 |
| chr7_-_149470297 | 0.13 |
ENST00000484747.1
|
ZNF467
|
zinc finger protein 467 |
| chr3_-_171489085 | 0.13 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr1_+_93544821 | 0.13 |
ENST00000370303.4
|
MTF2
|
metal response element binding transcription factor 2 |
| chr9_-_100954910 | 0.13 |
ENST00000375077.4
|
CORO2A
|
coronin, actin binding protein, 2A |
| chr10_-_32217717 | 0.13 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr5_-_176900610 | 0.13 |
ENST00000477391.2
ENST00000393565.1 ENST00000309007.5 |
DBN1
|
drebrin 1 |
| chr1_+_52521797 | 0.12 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr1_+_66458072 | 0.12 |
ENST00000423207.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr1_+_93913713 | 0.12 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr8_+_26240666 | 0.12 |
ENST00000523949.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chrX_+_135579670 | 0.12 |
ENST00000218364.4
|
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr7_+_11013491 | 0.12 |
ENST00000403050.3
ENST00000445996.2 |
PHF14
|
PHD finger protein 14 |
| chr1_+_201857798 | 0.12 |
ENST00000362011.6
|
SHISA4
|
shisa family member 4 |
| chr5_-_133510456 | 0.11 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr5_-_137071756 | 0.11 |
ENST00000394937.3
ENST00000309755.4 |
KLHL3
|
kelch-like family member 3 |
| chr16_+_57679859 | 0.11 |
ENST00000569494.1
ENST00000566169.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr19_+_16187816 | 0.11 |
ENST00000588410.1
|
TPM4
|
tropomyosin 4 |
| chr3_+_119187785 | 0.10 |
ENST00000295588.4
ENST00000476573.1 |
POGLUT1
|
protein O-glucosyltransferase 1 |
| chr21_+_35553045 | 0.10 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr11_+_111807863 | 0.10 |
ENST00000440460.2
|
DIXDC1
|
DIX domain containing 1 |
| chr17_+_53046096 | 0.09 |
ENST00000376352.2
ENST00000299341.4 ENST00000405898.1 ENST00000434978.2 ENST00000398391.2 |
STXBP4
|
syntaxin binding protein 4 |
| chr19_-_17375527 | 0.09 |
ENST00000431146.2
ENST00000594190.1 |
USHBP1
|
Usher syndrome 1C binding protein 1 |
| chrX_+_103173457 | 0.09 |
ENST00000419165.1
|
TMSB15B
|
thymosin beta 15B |
| chr1_-_26232951 | 0.09 |
ENST00000426559.2
ENST00000455785.2 |
STMN1
|
stathmin 1 |
| chr12_-_498415 | 0.09 |
ENST00000535014.1
ENST00000543507.1 ENST00000544760.1 |
KDM5A
|
lysine (K)-specific demethylase 5A |
| chr16_+_8814563 | 0.09 |
ENST00000425191.2
ENST00000569156.1 |
ABAT
|
4-aminobutyrate aminotransferase |
| chr19_-_55653259 | 0.09 |
ENST00000593194.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr1_+_52082751 | 0.09 |
ENST00000447887.1
ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9
|
oxysterol binding protein-like 9 |
| chr2_-_204400013 | 0.09 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr9_+_82267508 | 0.09 |
ENST00000490347.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr10_+_81466084 | 0.09 |
ENST00000342531.2
|
NUTM2B
|
NUT family member 2B |
| chr17_-_9939854 | 0.09 |
ENST00000584146.2
|
GAS7
|
growth arrest-specific 7 |
| chr18_+_74240756 | 0.09 |
ENST00000584910.1
ENST00000582452.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr13_-_45915221 | 0.08 |
ENST00000309246.5
ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1
|
tumor protein, translationally-controlled 1 |
| chr1_+_93544791 | 0.08 |
ENST00000545708.1
ENST00000540243.1 ENST00000370298.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr3_-_182698381 | 0.08 |
ENST00000292782.4
|
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr7_+_30067973 | 0.08 |
ENST00000258679.7
ENST00000449726.1 ENST00000396257.2 ENST00000396259.1 |
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chrX_+_103357202 | 0.08 |
ENST00000537356.3
|
ZCCHC18
|
zinc finger, CCHC domain containing 18 |
| chr6_+_32132360 | 0.08 |
ENST00000333845.6
ENST00000395512.1 ENST00000432129.1 |
EGFL8
|
EGF-like-domain, multiple 8 |
| chrX_-_38080077 | 0.08 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr1_-_154155675 | 0.08 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr1_-_104238912 | 0.08 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr2_+_228337079 | 0.08 |
ENST00000409315.1
ENST00000373671.3 ENST00000409171.1 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr12_+_98909520 | 0.08 |
ENST00000261210.5
|
TMPO
|
thymopoietin |
| chr8_+_133787586 | 0.07 |
ENST00000395379.1
ENST00000395386.2 ENST00000337920.4 |
PHF20L1
|
PHD finger protein 20-like 1 |
| chr22_+_50247449 | 0.07 |
ENST00000216268.5
|
ZBED4
|
zinc finger, BED-type containing 4 |
| chr12_-_54778471 | 0.07 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr1_+_23345943 | 0.07 |
ENST00000400181.4
ENST00000542151.1 |
KDM1A
|
lysine (K)-specific demethylase 1A |
| chr8_+_104427581 | 0.07 |
ENST00000521716.1
ENST00000521971.1 ENST00000519682.1 |
DCAF13
|
DDB1 and CUL4 associated factor 13 |
| chr22_+_21987005 | 0.07 |
ENST00000607942.1
ENST00000425975.1 ENST00000292779.3 |
CCDC116
|
coiled-coil domain containing 116 |
| chr16_+_57680043 | 0.07 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr7_-_105926058 | 0.07 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr1_-_40782938 | 0.07 |
ENST00000372736.3
ENST00000372748.3 |
COL9A2
|
collagen, type IX, alpha 2 |
| chr17_+_67498396 | 0.07 |
ENST00000588110.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr11_+_4116054 | 0.07 |
ENST00000423050.2
|
RRM1
|
ribonucleotide reductase M1 |
| chr1_-_6546001 | 0.07 |
ENST00000400913.1
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr13_-_101327028 | 0.06 |
ENST00000328767.5
ENST00000342624.5 ENST00000376234.3 ENST00000423847.1 |
TMTC4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr8_-_101117847 | 0.06 |
ENST00000523287.1
ENST00000519092.1 |
RGS22
|
regulator of G-protein signaling 22 |
| chr2_-_204399976 | 0.06 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr12_+_69004619 | 0.06 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr1_+_169075554 | 0.06 |
ENST00000367815.4
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr17_+_67498295 | 0.06 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr20_-_17662705 | 0.06 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr11_+_4116005 | 0.06 |
ENST00000300738.5
|
RRM1
|
ribonucleotide reductase M1 |
| chr1_-_27240455 | 0.06 |
ENST00000254227.3
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr6_+_97372645 | 0.06 |
ENST00000536676.1
ENST00000544166.1 |
KLHL32
|
kelch-like family member 32 |
| chr4_-_120548779 | 0.06 |
ENST00000264805.5
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific |
| chr7_-_107968921 | 0.05 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr8_-_101118185 | 0.05 |
ENST00000523437.1
|
RGS22
|
regulator of G-protein signaling 22 |
| chrX_+_100645812 | 0.05 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr12_+_98909260 | 0.05 |
ENST00000556029.1
|
TMPO
|
thymopoietin |
| chr1_+_23345930 | 0.05 |
ENST00000356634.3
|
KDM1A
|
lysine (K)-specific demethylase 1A |
| chrX_+_135579238 | 0.05 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr2_-_217560248 | 0.05 |
ENST00000233813.4
|
IGFBP5
|
insulin-like growth factor binding protein 5 |
| chr5_-_179050660 | 0.05 |
ENST00000519056.1
ENST00000506721.1 ENST00000503105.1 ENST00000504348.1 ENST00000508103.1 ENST00000510431.1 ENST00000515158.1 ENST00000393432.4 ENST00000442819.2 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr1_+_1266654 | 0.05 |
ENST00000339381.5
|
TAS1R3
|
taste receptor, type 1, member 3 |
| chr17_-_42100474 | 0.05 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr7_-_107968999 | 0.05 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr3_-_69591727 | 0.05 |
ENST00000459638.1
|
FRMD4B
|
FERM domain containing 4B |
| chr16_-_49890016 | 0.05 |
ENST00000563137.2
|
ZNF423
|
zinc finger protein 423 |
| chr6_+_127898312 | 0.05 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr3_-_57583052 | 0.05 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr6_-_3227877 | 0.05 |
ENST00000259818.7
|
TUBB2B
|
tubulin, beta 2B class IIb |
| chr8_-_134501937 | 0.05 |
ENST00000519924.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr6_+_97372596 | 0.04 |
ENST00000369261.4
|
KLHL32
|
kelch-like family member 32 |
| chr17_-_62308087 | 0.04 |
ENST00000583097.1
|
TEX2
|
testis expressed 2 |
| chr5_+_66124590 | 0.04 |
ENST00000490016.2
ENST00000403666.1 ENST00000450827.1 |
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_110620284 | 0.04 |
ENST00000529690.1
|
SYBU
|
syntabulin (syntaxin-interacting) |
| chr11_-_10830463 | 0.04 |
ENST00000527419.1
ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2 |
| chr3_+_51851612 | 0.04 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chrX_+_9433289 | 0.04 |
ENST00000422314.1
|
TBL1X
|
transducin (beta)-like 1X-linked |
| chr19_+_41768561 | 0.04 |
ENST00000599719.1
ENST00000601309.1 |
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr17_+_68101117 | 0.04 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_+_104198377 | 0.04 |
ENST00000370083.4
|
AMY1A
|
amylase, alpha 1A (salivary) |
| chr6_-_79944336 | 0.04 |
ENST00000344726.5
ENST00000275036.7 |
HMGN3
|
high mobility group nucleosomal binding domain 3 |
| chr16_+_77233294 | 0.04 |
ENST00000378644.4
|
SYCE1L
|
synaptonemal complex central element protein 1-like |
| chr2_-_55276320 | 0.04 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr10_+_69644404 | 0.04 |
ENST00000212015.6
|
SIRT1
|
sirtuin 1 |
| chr1_-_154155595 | 0.04 |
ENST00000328159.4
ENST00000368531.2 ENST00000323144.7 ENST00000368533.3 ENST00000341372.3 |
TPM3
|
tropomyosin 3 |
| chr11_-_18813353 | 0.04 |
ENST00000358540.2
ENST00000396171.4 ENST00000396167.2 |
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched) |
| chrX_+_21392873 | 0.04 |
ENST00000379510.3
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr22_+_38054721 | 0.04 |
ENST00000215904.6
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase |
| chr2_+_54350316 | 0.04 |
ENST00000606865.1
|
ACYP2
|
acylphosphatase 2, muscle type |
| chr2_-_37899323 | 0.04 |
ENST00000295324.3
ENST00000457889.1 |
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr1_+_181003067 | 0.04 |
ENST00000434571.2
ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1
|
major histocompatibility complex, class I-related |
| chr13_+_113656092 | 0.04 |
ENST00000397024.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr12_-_45270077 | 0.04 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr6_+_170190417 | 0.04 |
ENST00000420557.2
|
LINC00574
|
long intergenic non-protein coding RNA 574 |
| chr12_-_45270151 | 0.04 |
ENST00000429094.2
|
NELL2
|
NEL-like 2 (chicken) |
| chr11_-_79151695 | 0.04 |
ENST00000278550.7
|
TENM4
|
teneurin transmembrane protein 4 |
| chr11_-_62476965 | 0.03 |
ENST00000405837.1
ENST00000531524.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr19_+_58694396 | 0.03 |
ENST00000326804.4
ENST00000345813.3 ENST00000424679.2 |
ZNF274
|
zinc finger protein 274 |
| chr1_+_156119466 | 0.03 |
ENST00000414683.1
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr13_+_49684445 | 0.03 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr11_+_10772534 | 0.03 |
ENST00000361367.2
|
CTR9
|
CTR9, Paf1/RNA polymerase II complex component |
| chr1_-_26232522 | 0.03 |
ENST00000399728.1
|
STMN1
|
stathmin 1 |
| chr3_-_57583130 | 0.03 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr5_+_140213815 | 0.03 |
ENST00000525929.1
ENST00000378125.3 |
PCDHA7
|
protocadherin alpha 7 |
| chr12_-_95611149 | 0.03 |
ENST00000549499.1
ENST00000343958.4 ENST00000546711.1 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr2_+_102314161 | 0.03 |
ENST00000425019.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr7_-_128415844 | 0.03 |
ENST00000249389.2
|
OPN1SW
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr6_+_160183492 | 0.03 |
ENST00000541436.1
|
ACAT2
|
acetyl-CoA acetyltransferase 2 |
| chr3_-_57583185 | 0.03 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr14_+_76776957 | 0.03 |
ENST00000512784.1
|
ESRRB
|
estrogen-related receptor beta |
| chr11_+_128634589 | 0.03 |
ENST00000281428.8
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr3_-_129513259 | 0.03 |
ENST00000329333.5
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr12_+_69633372 | 0.03 |
ENST00000456847.3
ENST00000266679.8 |
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr17_-_9939935 | 0.03 |
ENST00000580043.1
|
GAS7
|
growth arrest-specific 7 |
| chr4_+_69962212 | 0.03 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr16_+_28874860 | 0.03 |
ENST00000545570.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr20_+_44036620 | 0.03 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr22_+_32439019 | 0.03 |
ENST00000266088.4
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr19_+_10765003 | 0.03 |
ENST00000407004.3
ENST00000589998.1 ENST00000589600.1 |
ILF3
|
interleukin enhancer binding factor 3, 90kDa |
| chrX_+_70503433 | 0.03 |
ENST00000276079.8
ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO
|
non-POU domain containing, octamer-binding |
| chr14_-_100772862 | 0.03 |
ENST00000359232.3
|
SLC25A29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr16_+_57680840 | 0.03 |
ENST00000563862.1
ENST00000564722.1 ENST00000569158.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr6_+_97372734 | 0.03 |
ENST00000539200.1
|
KLHL32
|
kelch-like family member 32 |
| chr19_-_5340730 | 0.03 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000349 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.2 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.1 | 0.5 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.2 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.1 | GO:2000584 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.7 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:0034721 | histone H3-K4 demethylation, trimethyl-H3-K4-specific(GO:0034721) |
| 0.0 | 0.2 | GO:1901189 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.2 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) response to fungicide(GO:0060992) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.2 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.0 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.2 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.1 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.0 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.5 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.2 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.0 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |