Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
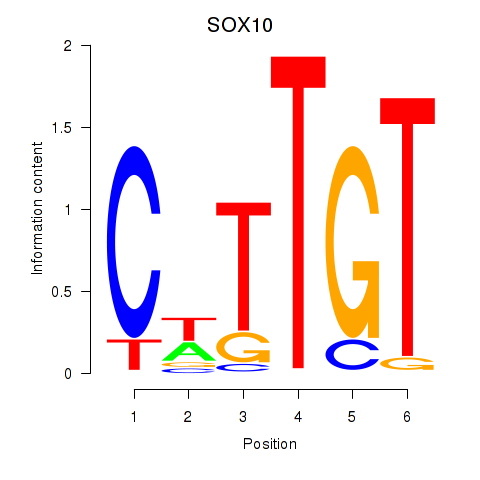
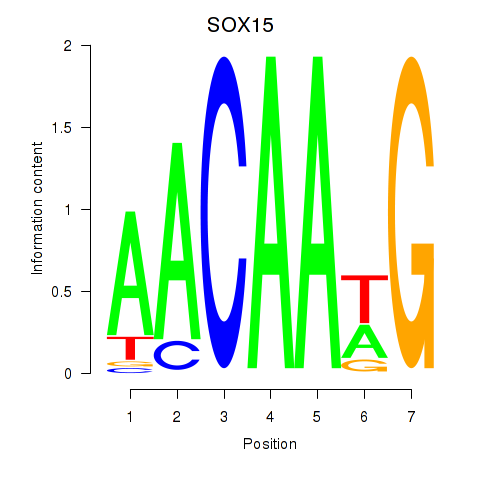
Results for SOX10_SOX15
Z-value: 0.18
Transcription factors associated with SOX10_SOX15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX10
|
ENSG00000100146.12 | SRY-box transcription factor 10 |
|
SOX15
|
ENSG00000129194.3 | SRY-box transcription factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX15 | hg19_v2_chr17_-_7493390_7493488 | -0.75 | 8.6e-02 | Click! |
Activity profile of SOX10_SOX15 motif
Sorted Z-values of SOX10_SOX15 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_51027838 | 0.22 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr7_+_23146271 | 0.21 |
ENST00000545771.1
|
KLHL7
|
kelch-like family member 7 |
| chr2_-_165697717 | 0.17 |
ENST00000439313.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chrX_-_45629661 | 0.17 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr4_+_95129061 | 0.14 |
ENST00000354268.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr1_-_65533390 | 0.13 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr12_+_95611569 | 0.13 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr15_-_52944231 | 0.13 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr1_-_26233423 | 0.12 |
ENST00000357865.2
|
STMN1
|
stathmin 1 |
| chr4_+_95128748 | 0.12 |
ENST00000359052.4
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_95128996 | 0.12 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr6_+_64282447 | 0.12 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr3_-_57583052 | 0.12 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr3_-_148939598 | 0.11 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr12_-_42631529 | 0.11 |
ENST00000548917.1
|
YAF2
|
YY1 associated factor 2 |
| chr10_+_22605374 | 0.10 |
ENST00000448361.1
|
COMMD3
|
COMM domain containing 3 |
| chrX_+_80457442 | 0.10 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr15_+_52311398 | 0.10 |
ENST00000261845.5
|
MAPK6
|
mitogen-activated protein kinase 6 |
| chr15_+_57210961 | 0.10 |
ENST00000557843.1
|
TCF12
|
transcription factor 12 |
| chr4_+_76649797 | 0.10 |
ENST00000538159.1
ENST00000514213.2 |
USO1
|
USO1 vesicle transport factor |
| chr15_-_45459704 | 0.09 |
ENST00000558039.1
|
CTD-2651B20.1
|
CTD-2651B20.1 |
| chr12_+_95611516 | 0.09 |
ENST00000436874.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr7_+_111846643 | 0.09 |
ENST00000361822.3
|
ZNF277
|
zinc finger protein 277 |
| chr7_+_111846741 | 0.09 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chr1_+_93645314 | 0.08 |
ENST00000343253.7
|
CCDC18
|
coiled-coil domain containing 18 |
| chr2_-_86564740 | 0.08 |
ENST00000540790.1
ENST00000428491.1 |
REEP1
|
receptor accessory protein 1 |
| chr5_-_39424961 | 0.08 |
ENST00000503513.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chrX_+_12993202 | 0.08 |
ENST00000451311.2
ENST00000380636.1 |
TMSB4X
|
thymosin beta 4, X-linked |
| chr6_-_64029879 | 0.08 |
ENST00000370658.5
ENST00000485906.2 ENST00000370657.4 |
LGSN
|
lengsin, lens protein with glutamine synthetase domain |
| chr8_-_42358742 | 0.08 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr6_+_119215308 | 0.08 |
ENST00000229595.5
|
ASF1A
|
anti-silencing function 1A histone chaperone |
| chr14_-_71107921 | 0.08 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr1_+_47799542 | 0.08 |
ENST00000471289.2
ENST00000450808.2 |
CMPK1
|
cytidine monophosphate (UMP-CMP) kinase 1, cytosolic |
| chr14_-_35099377 | 0.08 |
ENST00000362031.4
|
SNX6
|
sorting nexin 6 |
| chr14_-_35099315 | 0.07 |
ENST00000396526.3
ENST00000396534.3 ENST00000355110.5 ENST00000557265.1 |
SNX6
|
sorting nexin 6 |
| chr2_-_39348137 | 0.07 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr2_-_86564776 | 0.07 |
ENST00000165698.5
ENST00000541910.1 ENST00000535845.1 |
REEP1
|
receptor accessory protein 1 |
| chr6_+_87865262 | 0.07 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr4_+_76649753 | 0.07 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr12_+_95611536 | 0.07 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr6_+_122720681 | 0.07 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chrX_+_12993336 | 0.07 |
ENST00000380635.1
|
TMSB4X
|
thymosin beta 4, X-linked |
| chr11_-_46141338 | 0.07 |
ENST00000529782.1
ENST00000532010.1 ENST00000525438.1 ENST00000533757.1 ENST00000527782.1 |
PHF21A
|
PHD finger protein 21A |
| chr15_+_52043758 | 0.06 |
ENST00000249700.4
ENST00000539962.2 |
TMOD2
|
tropomodulin 2 (neuronal) |
| chr14_-_50583271 | 0.06 |
ENST00000395860.2
ENST00000395859.2 |
VCPKMT
|
valosin containing protein lysine (K) methyltransferase |
| chr14_+_54976603 | 0.06 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr6_+_56911476 | 0.06 |
ENST00000545356.1
|
KIAA1586
|
KIAA1586 |
| chr4_-_164534657 | 0.06 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr5_-_111092930 | 0.06 |
ENST00000257435.7
|
NREP
|
neuronal regeneration related protein |
| chr11_-_130184555 | 0.06 |
ENST00000525842.1
|
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr8_-_80993010 | 0.06 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr4_+_90033968 | 0.06 |
ENST00000317005.2
|
TIGD2
|
tigger transposable element derived 2 |
| chr5_-_65018834 | 0.06 |
ENST00000506816.1
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr8_+_107738240 | 0.06 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr7_+_23145884 | 0.05 |
ENST00000409689.1
ENST00000410047.1 |
KLHL7
|
kelch-like family member 7 |
| chr22_+_27053567 | 0.05 |
ENST00000449717.1
ENST00000453023.1 |
MIAT
|
myocardial infarction associated transcript (non-protein coding) |
| chr2_-_47572207 | 0.05 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr13_-_41593425 | 0.05 |
ENST00000239882.3
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr15_+_52043813 | 0.05 |
ENST00000435126.2
|
TMOD2
|
tropomodulin 2 (neuronal) |
| chr9_-_98268883 | 0.05 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr3_-_52719888 | 0.05 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr8_+_61822605 | 0.05 |
ENST00000526936.1
|
AC022182.1
|
AC022182.1 |
| chr2_+_169312725 | 0.05 |
ENST00000392687.4
|
CERS6
|
ceramide synthase 6 |
| chr15_+_65823092 | 0.05 |
ENST00000566074.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr11_-_111781554 | 0.05 |
ENST00000526167.1
ENST00000528961.1 |
CRYAB
|
crystallin, alpha B |
| chr12_-_95611149 | 0.05 |
ENST00000549499.1
ENST00000343958.4 ENST00000546711.1 |
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr5_-_111092873 | 0.05 |
ENST00000509025.1
ENST00000515855.1 |
NREP
|
neuronal regeneration related protein |
| chr2_+_70056762 | 0.05 |
ENST00000282570.3
|
GMCL1
|
germ cell-less, spermatogenesis associated 1 |
| chr5_-_65017921 | 0.05 |
ENST00000381007.4
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr17_+_53046096 | 0.05 |
ENST00000376352.2
ENST00000299341.4 ENST00000405898.1 ENST00000434978.2 ENST00000398391.2 |
STXBP4
|
syntaxin binding protein 4 |
| chr4_+_26323764 | 0.05 |
ENST00000514730.1
ENST00000507574.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr15_-_80695917 | 0.05 |
ENST00000559008.1
|
RP11-210M15.2
|
Uncharacterized protein |
| chr7_-_140178726 | 0.05 |
ENST00000480552.1
|
MKRN1
|
makorin ring finger protein 1 |
| chr14_-_35344093 | 0.05 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr11_-_102323489 | 0.05 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr7_-_15601595 | 0.05 |
ENST00000342526.3
|
AGMO
|
alkylglycerol monooxygenase |
| chr18_+_20494078 | 0.05 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr12_+_124997766 | 0.05 |
ENST00000543970.1
|
RP11-83B20.1
|
RP11-83B20.1 |
| chr3_-_148939835 | 0.05 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr16_+_53241854 | 0.04 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr2_+_32288657 | 0.04 |
ENST00000345662.1
|
SPAST
|
spastin |
| chr12_-_31477072 | 0.04 |
ENST00000454658.2
|
FAM60A
|
family with sequence similarity 60, member A |
| chr1_+_159141397 | 0.04 |
ENST00000368124.4
ENST00000368125.4 ENST00000416746.1 |
CADM3
|
cell adhesion molecule 3 |
| chr14_+_54976546 | 0.04 |
ENST00000216420.7
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr3_-_57583130 | 0.04 |
ENST00000303436.6
|
ARF4
|
ADP-ribosylation factor 4 |
| chr11_-_46848393 | 0.04 |
ENST00000526496.1
|
CKAP5
|
cytoskeleton associated protein 5 |
| chr2_+_120517174 | 0.04 |
ENST00000263708.2
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr9_-_74383302 | 0.04 |
ENST00000377066.5
|
TMEM2
|
transmembrane protein 2 |
| chr8_+_107738343 | 0.04 |
ENST00000521592.1
|
OXR1
|
oxidation resistance 1 |
| chrX_+_133507283 | 0.04 |
ENST00000370803.3
|
PHF6
|
PHD finger protein 6 |
| chrX_+_133507327 | 0.04 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr8_+_97597148 | 0.04 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr19_+_1452188 | 0.04 |
ENST00000587149.1
|
APC2
|
adenomatosis polyposis coli 2 |
| chr2_+_181845843 | 0.04 |
ENST00000602710.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr4_+_144303093 | 0.04 |
ENST00000505913.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr1_-_93645818 | 0.04 |
ENST00000370280.1
ENST00000479918.1 |
TMED5
|
transmembrane emp24 protein transport domain containing 5 |
| chr2_+_32288725 | 0.04 |
ENST00000315285.3
|
SPAST
|
spastin |
| chr12_+_69004619 | 0.04 |
ENST00000250559.9
ENST00000393436.5 ENST00000425247.2 ENST00000489473.2 ENST00000422358.2 ENST00000541167.1 ENST00000538283.1 ENST00000341355.5 ENST00000537460.1 ENST00000450214.2 ENST00000545270.1 ENST00000538980.1 ENST00000542018.1 ENST00000543393.1 |
RAP1B
|
RAP1B, member of RAS oncogene family |
| chr2_-_118943930 | 0.04 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr2_+_149402989 | 0.04 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr13_+_98795664 | 0.04 |
ENST00000376581.5
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr15_+_57211318 | 0.04 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr4_+_113152978 | 0.04 |
ENST00000309703.6
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein |
| chr12_+_31079652 | 0.04 |
ENST00000546076.1
ENST00000535215.1 ENST00000544427.1 ENST00000261177.9 |
TSPAN11
|
tetraspanin 11 |
| chr2_+_220309379 | 0.04 |
ENST00000451076.1
|
SPEG
|
SPEG complex locus |
| chr1_+_93913713 | 0.04 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr3_-_4927447 | 0.04 |
ENST00000449914.1
|
AC018816.3
|
Uncharacterized protein |
| chrX_+_78003204 | 0.03 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr9_-_123476612 | 0.03 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr4_+_26322987 | 0.03 |
ENST00000505958.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr15_+_78730531 | 0.03 |
ENST00000258886.8
|
IREB2
|
iron-responsive element binding protein 2 |
| chr21_+_30671690 | 0.03 |
ENST00000399921.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr21_-_16437126 | 0.03 |
ENST00000318948.4
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr9_-_119162885 | 0.03 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr1_+_166808692 | 0.03 |
ENST00000367876.4
|
POGK
|
pogo transposable element with KRAB domain |
| chr1_-_154155675 | 0.03 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr2_-_179343226 | 0.03 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr1_-_108231101 | 0.03 |
ENST00000544443.1
ENST00000415432.2 |
VAV3
|
vav 3 guanine nucleotide exchange factor |
| chrX_+_129305623 | 0.03 |
ENST00000257017.4
|
RAB33A
|
RAB33A, member RAS oncogene family |
| chr14_-_67981870 | 0.03 |
ENST00000555994.1
|
TMEM229B
|
transmembrane protein 229B |
| chr11_-_102323740 | 0.03 |
ENST00000398136.2
|
TMEM123
|
transmembrane protein 123 |
| chr2_-_55276320 | 0.03 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chrX_-_46187069 | 0.03 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr6_+_74405501 | 0.03 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr3_-_123339343 | 0.03 |
ENST00000578202.1
|
MYLK
|
myosin light chain kinase |
| chr14_+_96343100 | 0.03 |
ENST00000503525.2
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr1_+_228337553 | 0.03 |
ENST00000366714.2
|
GJC2
|
gap junction protein, gamma 2, 47kDa |
| chr7_-_76247617 | 0.03 |
ENST00000441393.1
|
POMZP3
|
POM121 and ZP3 fusion |
| chr5_+_65222438 | 0.03 |
ENST00000380938.2
|
ERBB2IP
|
erbb2 interacting protein |
| chr21_-_40720974 | 0.03 |
ENST00000380748.1
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr12_+_104359576 | 0.03 |
ENST00000392872.3
ENST00000436021.2 |
TDG
|
thymine-DNA glycosylase |
| chr7_+_29234101 | 0.03 |
ENST00000435288.2
|
CHN2
|
chimerin 2 |
| chr6_+_74405804 | 0.03 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr3_+_107241783 | 0.03 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr14_+_97263641 | 0.03 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr21_+_35553045 | 0.03 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr1_-_50889155 | 0.03 |
ENST00000404795.3
|
DMRTA2
|
DMRT-like family A2 |
| chr2_-_204399976 | 0.03 |
ENST00000457812.1
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr7_-_5821225 | 0.03 |
ENST00000416985.1
|
RNF216
|
ring finger protein 216 |
| chr4_+_154387480 | 0.03 |
ENST00000409663.3
ENST00000440693.1 ENST00000409959.3 |
KIAA0922
|
KIAA0922 |
| chrX_+_84498989 | 0.03 |
ENST00000395402.1
|
ZNF711
|
zinc finger protein 711 |
| chr12_-_90049878 | 0.03 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr14_+_59655369 | 0.03 |
ENST00000360909.3
ENST00000351081.1 ENST00000556135.1 |
DAAM1
|
dishevelled associated activator of morphogenesis 1 |
| chr3_-_52719912 | 0.03 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr13_+_78315295 | 0.03 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr12_-_102591604 | 0.03 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr1_-_115259337 | 0.03 |
ENST00000369535.4
|
NRAS
|
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr11_+_46402482 | 0.03 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr6_+_83777374 | 0.03 |
ENST00000349129.2
ENST00000237163.5 ENST00000536812.1 |
DOPEY1
|
dopey family member 1 |
| chr2_-_204400013 | 0.03 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr18_-_45456693 | 0.03 |
ENST00000587421.1
|
SMAD2
|
SMAD family member 2 |
| chr14_-_65409502 | 0.03 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr15_-_50647347 | 0.03 |
ENST00000220429.8
ENST00000429662.2 |
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr15_+_93426514 | 0.03 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr2_-_158182105 | 0.03 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr2_-_161350305 | 0.03 |
ENST00000348849.3
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1 |
| chr3_-_27525826 | 0.03 |
ENST00000454389.1
ENST00000440156.1 ENST00000437179.1 ENST00000446700.1 ENST00000455077.1 ENST00000435667.2 ENST00000388777.4 ENST00000425128.2 |
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr10_-_14646388 | 0.03 |
ENST00000468747.1
ENST00000378467.4 |
FAM107B
|
family with sequence similarity 107, member B |
| chr13_+_113656092 | 0.03 |
ENST00000397024.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr15_-_50647274 | 0.03 |
ENST00000543881.1
|
GABPB1
|
GA binding protein transcription factor, beta subunit 1 |
| chr9_+_131451480 | 0.03 |
ENST00000322030.8
|
SET
|
SET nuclear oncogene |
| chr1_-_91487013 | 0.03 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr17_+_78193443 | 0.03 |
ENST00000577155.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr6_-_139613269 | 0.03 |
ENST00000358430.3
|
TXLNB
|
taxilin beta |
| chr2_-_171571077 | 0.02 |
ENST00000409786.1
|
AC007405.2
|
long intergenic non-protein coding RNA 1124 |
| chr15_+_57210818 | 0.02 |
ENST00000438423.2
ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12
|
transcription factor 12 |
| chr15_-_52043722 | 0.02 |
ENST00000454181.2
|
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr13_+_49684445 | 0.02 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr8_+_21777159 | 0.02 |
ENST00000434536.1
ENST00000252512.9 |
XPO7
|
exportin 7 |
| chr5_+_78532003 | 0.02 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chrX_+_84499038 | 0.02 |
ENST00000373165.3
|
ZNF711
|
zinc finger protein 711 |
| chr10_-_15902449 | 0.02 |
ENST00000277632.3
|
FAM188A
|
family with sequence similarity 188, member A |
| chr6_+_27114861 | 0.02 |
ENST00000377459.1
|
HIST1H2AH
|
histone cluster 1, H2ah |
| chr12_-_76953284 | 0.02 |
ENST00000547544.1
ENST00000393249.2 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr6_+_17281802 | 0.02 |
ENST00000509686.1
|
RBM24
|
RNA binding motif protein 24 |
| chr3_+_107241882 | 0.02 |
ENST00000416476.2
|
BBX
|
bobby sox homolog (Drosophila) |
| chr15_-_67439270 | 0.02 |
ENST00000558463.1
|
RP11-342M21.2
|
Uncharacterized protein |
| chr4_+_41614909 | 0.02 |
ENST00000509454.1
ENST00000396595.3 ENST00000381753.4 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr1_-_182360498 | 0.02 |
ENST00000417584.2
|
GLUL
|
glutamate-ammonia ligase |
| chr10_+_114709999 | 0.02 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_-_71551868 | 0.02 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr2_+_153191706 | 0.02 |
ENST00000288670.9
|
FMNL2
|
formin-like 2 |
| chr2_+_24150180 | 0.02 |
ENST00000404924.1
|
UBXN2A
|
UBX domain protein 2A |
| chr3_-_141747950 | 0.02 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_-_53046058 | 0.02 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr7_+_74379083 | 0.02 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr12_-_90049828 | 0.02 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr5_-_114515734 | 0.02 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr16_+_23765948 | 0.02 |
ENST00000300113.2
|
CHP2
|
calcineurin-like EF-hand protein 2 |
| chr14_-_31495569 | 0.02 |
ENST00000357479.5
ENST00000355683.5 |
STRN3
|
striatin, calmodulin binding protein 3 |
| chr1_+_66797687 | 0.02 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr21_-_40720995 | 0.02 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr13_-_24007815 | 0.02 |
ENST00000382298.3
|
SACS
|
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr6_+_56954867 | 0.02 |
ENST00000370708.4
ENST00000370702.1 |
ZNF451
|
zinc finger protein 451 |
| chr12_-_45270077 | 0.02 |
ENST00000551601.1
ENST00000549027.1 ENST00000452445.2 |
NELL2
|
NEL-like 2 (chicken) |
| chr12_+_26111823 | 0.02 |
ENST00000381352.3
ENST00000535907.1 ENST00000405154.2 |
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr6_+_151358048 | 0.02 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr1_+_244998602 | 0.02 |
ENST00000411948.2
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr22_+_31742875 | 0.02 |
ENST00000504184.2
|
AC005003.1
|
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chr11_-_111781454 | 0.02 |
ENST00000533280.1
|
CRYAB
|
crystallin, alpha B |
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX10_SOX15
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.0 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.0 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.0 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:1901297 | positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.0 | GO:0008623 | CHRAC(GO:0008623) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.0 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |