Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
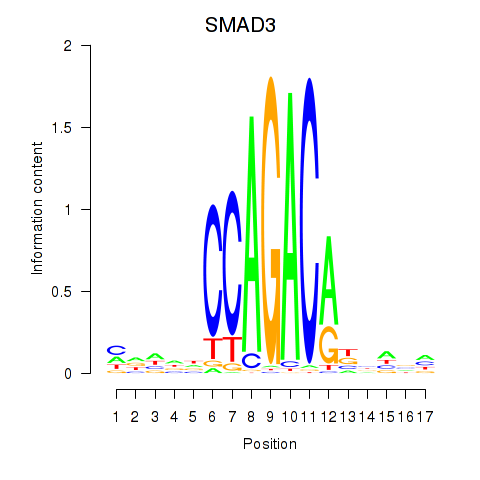
Results for SMAD3
Z-value: 0.24
Transcription factors associated with SMAD3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD3
|
ENSG00000166949.11 | SMAD family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD3 | hg19_v2_chr15_+_67358163_67358192 | 0.65 | 1.6e-01 | Click! |
Activity profile of SMAD3 motif
Sorted Z-values of SMAD3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_27775694 | 0.41 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chr6_-_27806117 | 0.14 |
ENST00000330180.2
|
HIST1H2AK
|
histone cluster 1, H2ak |
| chr22_+_20877924 | 0.14 |
ENST00000445189.1
|
MED15
|
mediator complex subunit 15 |
| chr19_-_36304201 | 0.13 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr6_-_26124138 | 0.12 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr4_-_90759440 | 0.12 |
ENST00000336904.3
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr5_+_127039075 | 0.12 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr18_+_13612613 | 0.12 |
ENST00000586765.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chr14_-_100625932 | 0.11 |
ENST00000553834.1
|
DEGS2
|
delta(4)-desaturase, sphingolipid 2 |
| chrX_+_49832231 | 0.11 |
ENST00000376108.3
|
CLCN5
|
chloride channel, voltage-sensitive 5 |
| chr16_+_19098178 | 0.11 |
ENST00000568032.1
|
RP11-626G11.4
|
RP11-626G11.4 |
| chr20_-_43280361 | 0.10 |
ENST00000372874.4
|
ADA
|
adenosine deaminase |
| chr11_+_72983246 | 0.10 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr19_+_51728316 | 0.10 |
ENST00000436584.2
ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33
|
CD33 molecule |
| chr4_-_177116772 | 0.10 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr6_+_27791862 | 0.10 |
ENST00000355057.1
|
HIST1H4J
|
histone cluster 1, H4j |
| chr15_-_82338460 | 0.09 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr1_+_202385953 | 0.09 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr14_+_93389425 | 0.09 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr6_+_26273144 | 0.09 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr1_+_39796810 | 0.09 |
ENST00000289893.4
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr19_-_35166604 | 0.09 |
ENST00000601241.1
|
SCGB2B2
|
secretoglobin, family 2B, member 2 |
| chr4_+_186990298 | 0.09 |
ENST00000296795.3
ENST00000513189.1 |
TLR3
|
toll-like receptor 3 |
| chr11_+_113258495 | 0.09 |
ENST00000303941.3
|
ANKK1
|
ankyrin repeat and kinase domain containing 1 |
| chr11_+_117070037 | 0.09 |
ENST00000392951.4
ENST00000525531.1 ENST00000278968.6 |
TAGLN
|
transgelin |
| chr6_-_27782548 | 0.08 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr9_-_14322319 | 0.08 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr19_-_13947099 | 0.08 |
ENST00000587762.1
|
MIR24-2
|
microRNA 24-2 |
| chr1_-_9129735 | 0.08 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr6_+_27806319 | 0.08 |
ENST00000606613.1
ENST00000396980.3 |
HIST1H2BN
|
histone cluster 1, H2bn |
| chr16_-_84220633 | 0.08 |
ENST00000566732.1
ENST00000561955.1 ENST00000564454.1 ENST00000341690.6 ENST00000541676.1 ENST00000570117.1 ENST00000564345.1 |
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr16_+_14280742 | 0.08 |
ENST00000341243.5
|
MKL2
|
MKL/myocardin-like 2 |
| chr1_+_200863949 | 0.08 |
ENST00000413687.2
|
C1orf106
|
chromosome 1 open reading frame 106 |
| chr2_-_220110111 | 0.07 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chr15_-_83316254 | 0.07 |
ENST00000567678.1
ENST00000450751.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr20_-_43280325 | 0.07 |
ENST00000537820.1
|
ADA
|
adenosine deaminase |
| chr4_+_40337340 | 0.07 |
ENST00000310169.2
|
CHRNA9
|
cholinergic receptor, nicotinic, alpha 9 (neuronal) |
| chr6_+_116937636 | 0.07 |
ENST00000368581.4
ENST00000229554.5 ENST00000368580.4 |
RSPH4A
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr11_-_61101247 | 0.07 |
ENST00000543627.1
|
DDB1
|
damage-specific DNA binding protein 1, 127kDa |
| chr7_+_139528952 | 0.07 |
ENST00000416849.2
ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr11_+_57531292 | 0.07 |
ENST00000524579.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr10_-_106098162 | 0.07 |
ENST00000337478.1
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr7_+_139529040 | 0.07 |
ENST00000455353.1
ENST00000458722.1 ENST00000411653.1 |
TBXAS1
|
thromboxane A synthase 1 (platelet) |
| chr7_-_55620433 | 0.06 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr10_-_105437909 | 0.06 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr17_-_34195862 | 0.06 |
ENST00000592980.1
ENST00000587626.1 |
C17orf66
|
chromosome 17 open reading frame 66 |
| chr16_-_31076332 | 0.06 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr6_-_11807277 | 0.06 |
ENST00000379415.2
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr16_+_699319 | 0.06 |
ENST00000549091.1
ENST00000293879.4 |
WDR90
|
WD repeat domain 90 |
| chr14_+_91581011 | 0.06 |
ENST00000523894.1
ENST00000522322.1 ENST00000523771.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr8_+_38586068 | 0.06 |
ENST00000443286.2
ENST00000520340.1 ENST00000518415.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr4_+_69917078 | 0.06 |
ENST00000502942.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr16_-_11723066 | 0.06 |
ENST00000576036.1
|
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr16_-_84220604 | 0.06 |
ENST00000567759.1
|
TAF1C
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, C, 110kDa |
| chr16_+_14280564 | 0.05 |
ENST00000572567.1
|
MKL2
|
MKL/myocardin-like 2 |
| chr19_-_42724261 | 0.05 |
ENST00000595337.1
|
DEDD2
|
death effector domain containing 2 |
| chr14_+_91580777 | 0.05 |
ENST00000525393.2
ENST00000428926.2 ENST00000517362.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr14_+_102786063 | 0.05 |
ENST00000442396.2
ENST00000262236.5 |
ZNF839
|
zinc finger protein 839 |
| chr7_-_22862406 | 0.05 |
ENST00000372879.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr19_-_42006539 | 0.05 |
ENST00000597702.1
ENST00000588495.1 ENST00000595837.1 ENST00000594315.1 |
AC011526.1
|
AC011526.1 |
| chr3_+_51741072 | 0.05 |
ENST00000395052.3
|
GRM2
|
glutamate receptor, metabotropic 2 |
| chr16_-_30773372 | 0.05 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr17_+_30813576 | 0.05 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr1_-_156399184 | 0.05 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr16_-_31076273 | 0.05 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr17_-_73874654 | 0.05 |
ENST00000254816.2
|
TRIM47
|
tripartite motif containing 47 |
| chr1_-_36906474 | 0.05 |
ENST00000433045.2
|
OSCP1
|
organic solute carrier partner 1 |
| chr9_+_5890802 | 0.05 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr19_+_42724423 | 0.05 |
ENST00000301215.3
ENST00000597945.1 |
ZNF526
|
zinc finger protein 526 |
| chr17_-_982198 | 0.05 |
ENST00000571945.1
ENST00000536794.2 |
ABR
|
active BCR-related |
| chr3_+_12838161 | 0.05 |
ENST00000456430.2
|
CAND2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr1_-_9129631 | 0.04 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr14_+_96858454 | 0.04 |
ENST00000555570.1
|
AK7
|
adenylate kinase 7 |
| chr11_+_46383121 | 0.04 |
ENST00000454345.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr22_-_36635225 | 0.04 |
ENST00000529194.1
|
APOL2
|
apolipoprotein L, 2 |
| chrX_-_62571220 | 0.04 |
ENST00000374884.2
|
SPIN4
|
spindlin family, member 4 |
| chrX_-_62571187 | 0.04 |
ENST00000335144.3
|
SPIN4
|
spindlin family, member 4 |
| chr6_-_27100529 | 0.04 |
ENST00000607124.1
ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ
|
histone cluster 1, H2bj |
| chr16_+_56703737 | 0.04 |
ENST00000569155.1
|
MT1H
|
metallothionein 1H |
| chr18_+_43753500 | 0.04 |
ENST00000587591.1
ENST00000588730.1 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr22_-_41677625 | 0.04 |
ENST00000452543.1
ENST00000418067.1 |
RANGAP1
|
Ran GTPase activating protein 1 |
| chr1_+_149804218 | 0.04 |
ENST00000610125.1
|
HIST2H4A
|
histone cluster 2, H4a |
| chr4_+_2814011 | 0.04 |
ENST00000502260.1
ENST00000435136.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr1_-_8877692 | 0.04 |
ENST00000400908.2
|
RERE
|
arginine-glutamic acid dipeptide (RE) repeats |
| chr18_+_44526744 | 0.04 |
ENST00000585469.1
|
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr9_+_37486005 | 0.04 |
ENST00000377792.3
|
POLR1E
|
polymerase (RNA) I polypeptide E, 53kDa |
| chr2_-_208031943 | 0.04 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr12_+_49212514 | 0.04 |
ENST00000301050.2
ENST00000548279.1 ENST00000547230.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr11_+_62538775 | 0.04 |
ENST00000294168.3
ENST00000526261.1 |
TAF6L
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
| chr22_+_41956767 | 0.04 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr9_+_37485932 | 0.04 |
ENST00000377798.4
ENST00000442009.2 |
POLR1E
|
polymerase (RNA) I polypeptide E, 53kDa |
| chr5_-_180242534 | 0.03 |
ENST00000333055.3
ENST00000513431.1 |
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr17_-_43487780 | 0.03 |
ENST00000532038.1
ENST00000528677.1 |
ARHGAP27
|
Rho GTPase activating protein 27 |
| chr14_+_91580708 | 0.03 |
ENST00000518868.1
|
C14orf159
|
chromosome 14 open reading frame 159 |
| chr1_+_52682052 | 0.03 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr1_+_59762642 | 0.03 |
ENST00000371218.4
ENST00000303721.7 |
FGGY
|
FGGY carbohydrate kinase domain containing |
| chr22_-_36635598 | 0.03 |
ENST00000454728.1
|
APOL2
|
apolipoprotein L, 2 |
| chr19_-_41196458 | 0.03 |
ENST00000598779.1
|
NUMBL
|
numb homolog (Drosophila)-like |
| chr3_+_126243126 | 0.03 |
ENST00000319340.2
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13 |
| chr6_-_41888843 | 0.03 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chrX_-_2847366 | 0.03 |
ENST00000381154.1
|
ARSD
|
arylsulfatase D |
| chr6_+_30850697 | 0.03 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr14_+_91580357 | 0.03 |
ENST00000298858.4
ENST00000521081.1 ENST00000520328.1 ENST00000256324.10 ENST00000524232.1 ENST00000522170.1 ENST00000519950.1 ENST00000523879.1 ENST00000521077.2 ENST00000518665.2 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr16_+_30773636 | 0.03 |
ENST00000402121.3
ENST00000565995.1 ENST00000563683.1 ENST00000357890.5 ENST00000565931.1 |
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr1_-_44482979 | 0.03 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr10_-_24911746 | 0.03 |
ENST00000320481.6
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr7_-_22862448 | 0.03 |
ENST00000358435.4
|
TOMM7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr12_+_119616447 | 0.03 |
ENST00000281938.2
|
HSPB8
|
heat shock 22kDa protein 8 |
| chr14_+_91580732 | 0.03 |
ENST00000519019.1
ENST00000523816.1 ENST00000517518.1 |
C14orf159
|
chromosome 14 open reading frame 159 |
| chr5_+_140529630 | 0.02 |
ENST00000543635.1
|
PCDHB6
|
protocadherin beta 6 |
| chr2_+_220110177 | 0.02 |
ENST00000409638.3
ENST00000396738.2 ENST00000409516.3 |
STK16
|
serine/threonine kinase 16 |
| chr20_+_24449821 | 0.02 |
ENST00000376862.3
|
SYNDIG1
|
synapse differentiation inducing 1 |
| chr18_+_29598335 | 0.02 |
ENST00000217740.3
|
RNF125
|
ring finger protein 125, E3 ubiquitin protein ligase |
| chr22_-_36635563 | 0.02 |
ENST00000451256.2
|
APOL2
|
apolipoprotein L, 2 |
| chr2_-_75788424 | 0.02 |
ENST00000410071.1
|
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr7_+_141490017 | 0.02 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr20_+_44036900 | 0.02 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_+_35211511 | 0.02 |
ENST00000524922.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr5_-_79950775 | 0.02 |
ENST00000439211.2
|
DHFR
|
dihydrofolate reductase |
| chr12_-_9913489 | 0.02 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr1_+_201159914 | 0.02 |
ENST00000335211.4
ENST00000451870.2 ENST00000295591.8 |
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1 |
| chr4_+_2813946 | 0.02 |
ENST00000442312.2
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr18_+_77623668 | 0.02 |
ENST00000316249.3
|
KCNG2
|
potassium voltage-gated channel, subfamily G, member 2 |
| chr16_-_3306587 | 0.02 |
ENST00000541159.1
ENST00000536379.1 ENST00000219596.1 ENST00000339854.4 |
MEFV
|
Mediterranean fever |
| chr16_-_58328923 | 0.02 |
ENST00000567164.1
ENST00000219301.4 ENST00000569727.1 |
PRSS54
|
protease, serine, 54 |
| chr13_-_114898016 | 0.02 |
ENST00000542651.1
ENST00000334062.7 |
RASA3
|
RAS p21 protein activator 3 |
| chr15_-_83316087 | 0.02 |
ENST00000568757.1
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr2_-_220110187 | 0.02 |
ENST00000295759.7
ENST00000392089.2 |
GLB1L
|
galactosidase, beta 1-like |
| chr3_-_14220068 | 0.02 |
ENST00000449060.2
ENST00000511155.1 |
XPC
|
xeroderma pigmentosum, complementation group C |
| chr8_+_22250334 | 0.02 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr15_+_63889552 | 0.02 |
ENST00000360587.2
|
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr12_+_14927270 | 0.02 |
ENST00000544848.1
|
H2AFJ
|
H2A histone family, member J |
| chr11_+_66406149 | 0.01 |
ENST00000578778.1
ENST00000483858.1 ENST00000398692.4 ENST00000510173.2 ENST00000506523.2 ENST00000530235.1 ENST00000532968.1 |
RBM4
|
RNA binding motif protein 4 |
| chr4_-_186696515 | 0.01 |
ENST00000456596.1
ENST00000414724.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr7_+_39125365 | 0.01 |
ENST00000559001.1
ENST00000464276.2 |
POU6F2
|
POU class 6 homeobox 2 |
| chr4_-_186696425 | 0.01 |
ENST00000430503.1
ENST00000319454.6 ENST00000450341.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_+_140566 | 0.01 |
ENST00000502646.1
|
PLEKHG4B
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B |
| chr8_+_38585704 | 0.01 |
ENST00000519416.1
ENST00000520615.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr19_-_50143452 | 0.01 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr22_-_46644182 | 0.01 |
ENST00000404583.1
ENST00000404744.1 |
CDPF1
|
cysteine-rich, DPF motif domain containing 1 |
| chr16_-_57798253 | 0.01 |
ENST00000565270.1
|
KIFC3
|
kinesin family member C3 |
| chr1_-_89357179 | 0.01 |
ENST00000448623.1
ENST00000418217.1 ENST00000370500.5 |
GTF2B
|
general transcription factor IIB |
| chr16_-_58328870 | 0.01 |
ENST00000543437.1
|
PRSS54
|
protease, serine, 54 |
| chr16_+_618837 | 0.01 |
ENST00000409439.2
|
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr2_+_28615669 | 0.01 |
ENST00000379619.1
ENST00000264716.4 |
FOSL2
|
FOS-like antigen 2 |
| chr16_+_57847684 | 0.01 |
ENST00000335616.2
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr11_-_65150103 | 0.01 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chrX_-_46618490 | 0.01 |
ENST00000328306.4
|
SLC9A7
|
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr11_-_72145426 | 0.01 |
ENST00000535990.1
ENST00000437826.2 ENST00000340729.5 |
CLPB
|
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr4_-_186696561 | 0.01 |
ENST00000445115.1
ENST00000451701.1 ENST00000457247.1 ENST00000435480.1 ENST00000425679.1 ENST00000457934.1 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr5_-_180242576 | 0.01 |
ENST00000514438.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr4_-_38666430 | 0.01 |
ENST00000436901.1
|
AC021860.1
|
Uncharacterized protein |
| chr8_+_145065705 | 0.01 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr1_-_11042094 | 0.01 |
ENST00000377004.4
ENST00000377008.4 |
C1orf127
|
chromosome 1 open reading frame 127 |
| chr15_+_22736484 | 0.01 |
ENST00000560659.2
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chr20_-_30311703 | 0.01 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr8_+_128747661 | 0.01 |
ENST00000259523.6
|
MYC
|
v-myc avian myelocytomatosis viral oncogene homolog |
| chr6_-_66417107 | 0.01 |
ENST00000370621.3
ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS
|
eyes shut homolog (Drosophila) |
| chr1_-_160040038 | 0.01 |
ENST00000368089.3
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr20_+_58251716 | 0.01 |
ENST00000355648.4
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr10_-_90712520 | 0.01 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr5_+_55147205 | 0.00 |
ENST00000396836.2
ENST00000396834.1 ENST00000447346.2 ENST00000359040.5 |
IL31RA
|
interleukin 31 receptor A |
| chr10_+_129796390 | 0.00 |
ENST00000455661.1
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr11_-_3147835 | 0.00 |
ENST00000525498.1
|
OSBPL5
|
oxysterol binding protein-like 5 |
| chr10_+_94451574 | 0.00 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr17_+_36452989 | 0.00 |
ENST00000312513.5
ENST00000582535.1 |
MRPL45
|
mitochondrial ribosomal protein L45 |
| chr1_+_209602771 | 0.00 |
ENST00000440276.1
|
MIR205HG
|
MIR205 host gene (non-protein coding) |
| chr6_+_30851205 | 0.00 |
ENST00000515881.1
|
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr15_-_43785303 | 0.00 |
ENST00000382039.3
ENST00000450115.2 ENST00000382044.4 |
TP53BP1
|
tumor protein p53 binding protein 1 |
| chr6_-_99842041 | 0.00 |
ENST00000254759.3
ENST00000369242.1 |
COQ3
|
coenzyme Q3 methyltransferase |
| chr14_-_65409502 | 0.00 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr16_+_30484054 | 0.00 |
ENST00000564118.1
ENST00000454514.2 ENST00000433423.2 |
ITGAL
|
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
| chr17_-_56621665 | 0.00 |
ENST00000321691.3
|
C17orf47
|
chromosome 17 open reading frame 47 |
| chr19_+_34850385 | 0.00 |
ENST00000587521.2
ENST00000587384.1 ENST00000592277.1 |
GPI
|
glucose-6-phosphate isomerase |
| chr9_+_100069933 | 0.00 |
ENST00000529487.1
|
CCDC180
|
coiled-coil domain containing 180 |
| chr11_+_66406088 | 0.00 |
ENST00000310092.7
ENST00000396053.4 ENST00000408993.2 |
RBM4
|
RNA binding motif protein 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0034344 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0001189 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.0 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:1990539 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.0 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.2 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.0 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.0 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |