Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
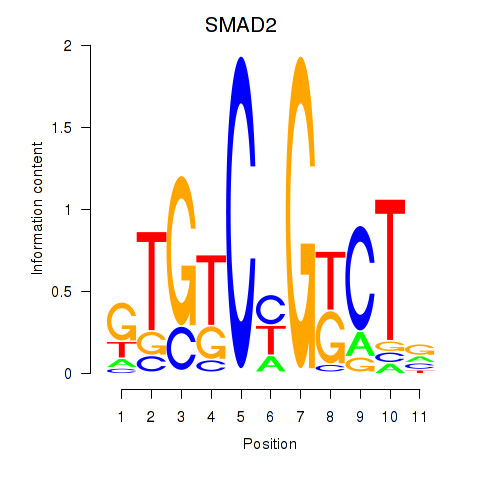
Results for SMAD2
Z-value: 1.13
Transcription factors associated with SMAD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD2
|
ENSG00000175387.11 | SMAD family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMAD2 | hg19_v2_chr18_-_45456930_45456970 | -0.89 | 1.8e-02 | Click! |
Activity profile of SMAD2 motif
Sorted Z-values of SMAD2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_20715416 | 1.58 |
ENST00000580153.1
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr7_-_155601766 | 1.28 |
ENST00000430104.1
|
SHH
|
sonic hedgehog |
| chr19_-_49522727 | 1.15 |
ENST00000600007.1
|
CTB-60B18.10
|
CTB-60B18.10 |
| chr5_-_151066514 | 0.94 |
ENST00000538026.1
ENST00000522348.1 ENST00000521569.1 |
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr12_-_53207842 | 0.91 |
ENST00000458244.2
|
KRT4
|
keratin 4 |
| chr6_-_152489484 | 0.67 |
ENST00000354674.4
ENST00000539504.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr12_-_6740802 | 0.66 |
ENST00000431922.1
|
LPAR5
|
lysophosphatidic acid receptor 5 |
| chr2_-_228028829 | 0.56 |
ENST00000396625.3
ENST00000329662.7 |
COL4A4
|
collagen, type IV, alpha 4 |
| chr15_-_82338460 | 0.49 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr17_+_73629500 | 0.49 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr12_-_120241187 | 0.49 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr11_-_75379479 | 0.44 |
ENST00000434603.2
|
MAP6
|
microtubule-associated protein 6 |
| chr8_-_130587237 | 0.40 |
ENST00000520048.1
|
CCDC26
|
coiled-coil domain containing 26 |
| chr7_+_69064566 | 0.39 |
ENST00000403018.2
|
AUTS2
|
autism susceptibility candidate 2 |
| chr1_+_33722080 | 0.38 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr1_-_149783914 | 0.37 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chr3_+_111393659 | 0.37 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr3_+_195447738 | 0.36 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr11_-_117166201 | 0.35 |
ENST00000510915.1
|
BACE1
|
beta-site APP-cleaving enzyme 1 |
| chr14_+_70078303 | 0.35 |
ENST00000342745.4
|
KIAA0247
|
KIAA0247 |
| chr17_-_7297833 | 0.35 |
ENST00000571802.1
ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr10_+_80027105 | 0.34 |
ENST00000461034.1
ENST00000476909.1 ENST00000459633.1 |
LINC00595
|
long intergenic non-protein coding RNA 595 |
| chr5_+_14143728 | 0.34 |
ENST00000344204.4
ENST00000537187.1 |
TRIO
|
trio Rho guanine nucleotide exchange factor |
| chr8_+_95907993 | 0.34 |
ENST00000523378.1
|
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr12_-_108991778 | 0.33 |
ENST00000549447.1
|
TMEM119
|
transmembrane protein 119 |
| chr19_+_50094866 | 0.32 |
ENST00000418929.2
|
PRR12
|
proline rich 12 |
| chr20_+_1875942 | 0.32 |
ENST00000358771.4
|
SIRPA
|
signal-regulatory protein alpha |
| chr12_+_122516626 | 0.32 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr17_+_65375082 | 0.32 |
ENST00000584471.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr16_+_28889801 | 0.32 |
ENST00000395503.4
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr9_+_140145713 | 0.31 |
ENST00000388931.3
ENST00000412566.1 |
C9orf173
|
chromosome 9 open reading frame 173 |
| chr19_-_51587502 | 0.31 |
ENST00000156499.2
ENST00000391802.1 |
KLK14
|
kallikrein-related peptidase 14 |
| chr17_-_3571934 | 0.30 |
ENST00000225525.3
|
TAX1BP3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr11_+_64808675 | 0.30 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr19_+_50148087 | 0.30 |
ENST00000601038.1
ENST00000595242.1 |
SCAF1
|
SR-related CTD-associated factor 1 |
| chr19_+_10527449 | 0.30 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr16_-_87417351 | 0.29 |
ENST00000311635.7
|
FBXO31
|
F-box protein 31 |
| chr1_-_44482979 | 0.29 |
ENST00000360584.2
ENST00000357730.2 ENST00000528803.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr16_-_70719925 | 0.29 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr19_-_3551043 | 0.29 |
ENST00000589995.1
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr20_-_56284816 | 0.28 |
ENST00000395819.3
ENST00000341744.3 |
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr6_-_143771799 | 0.28 |
ENST00000237283.8
|
ADAT2
|
adenosine deaminase, tRNA-specific 2 |
| chrX_+_48660107 | 0.28 |
ENST00000376643.2
|
HDAC6
|
histone deacetylase 6 |
| chr19_-_6481776 | 0.27 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr5_-_172755056 | 0.27 |
ENST00000520648.1
|
STC2
|
stanniocalcin 2 |
| chr19_-_55580838 | 0.27 |
ENST00000396247.3
ENST00000586945.1 ENST00000587026.1 |
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr1_-_1243252 | 0.27 |
ENST00000353662.3
|
ACAP3
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
| chr17_-_40761375 | 0.27 |
ENST00000543197.1
ENST00000309428.5 |
FAM134C
|
family with sequence similarity 134, member C |
| chr16_+_28889703 | 0.25 |
ENST00000357084.3
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr22_-_43567750 | 0.25 |
ENST00000494035.1
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr22_+_22673051 | 0.25 |
ENST00000390289.2
|
IGLV5-52
|
immunoglobulin lambda variable 5-52 |
| chr21_-_36259445 | 0.25 |
ENST00000399240.1
|
RUNX1
|
runt-related transcription factor 1 |
| chr3_-_195603566 | 0.24 |
ENST00000424563.1
ENST00000411741.1 |
TNK2
|
tyrosine kinase, non-receptor, 2 |
| chrX_+_48660287 | 0.24 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr10_+_105314881 | 0.24 |
ENST00000437579.1
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr1_-_44497118 | 0.23 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr20_+_44098385 | 0.23 |
ENST00000217425.5
ENST00000339946.3 |
WFDC2
|
WAP four-disulfide core domain 2 |
| chr1_-_149908710 | 0.22 |
ENST00000439741.2
ENST00000361405.6 ENST00000406732.3 |
MTMR11
|
myotubularin related protein 11 |
| chr11_-_118789613 | 0.22 |
ENST00000532899.1
|
BCL9L
|
B-cell CLL/lymphoma 9-like |
| chr16_+_31483374 | 0.22 |
ENST00000394863.3
|
TGFB1I1
|
transforming growth factor beta 1 induced transcript 1 |
| chr11_+_2923423 | 0.21 |
ENST00000312221.5
|
SLC22A18
|
solute carrier family 22, member 18 |
| chr7_+_99905325 | 0.21 |
ENST00000332397.6
ENST00000437326.2 |
SPDYE3
|
speedy/RINGO cell cycle regulator family member E3 |
| chr3_+_50316458 | 0.21 |
ENST00000316436.3
|
LSMEM2
|
leucine-rich single-pass membrane protein 2 |
| chr22_-_37823468 | 0.20 |
ENST00000402918.2
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr1_-_935491 | 0.20 |
ENST00000304952.6
|
HES4
|
hes family bHLH transcription factor 4 |
| chr6_+_32938665 | 0.20 |
ENST00000374831.4
ENST00000395289.2 |
BRD2
|
bromodomain containing 2 |
| chr11_+_75273101 | 0.20 |
ENST00000533603.1
ENST00000358171.3 ENST00000526242.1 |
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr17_+_30593195 | 0.20 |
ENST00000431505.2
ENST00000269051.4 ENST00000538145.1 |
RHBDL3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr8_+_144373550 | 0.20 |
ENST00000330143.3
ENST00000521537.1 ENST00000518432.1 ENST00000520333.1 |
ZNF696
|
zinc finger protein 696 |
| chr8_+_11561660 | 0.20 |
ENST00000526716.1
ENST00000335135.4 ENST00000528027.1 |
GATA4
|
GATA binding protein 4 |
| chr11_-_3859089 | 0.20 |
ENST00000396979.1
|
RHOG
|
ras homolog family member G |
| chr12_+_47473369 | 0.19 |
ENST00000546455.1
|
PCED1B
|
PC-esterase domain containing 1B |
| chr12_-_6798523 | 0.19 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr19_+_56186557 | 0.19 |
ENST00000270460.6
|
EPN1
|
epsin 1 |
| chr1_+_44412577 | 0.19 |
ENST00000372343.3
|
IPO13
|
importin 13 |
| chr1_+_174933899 | 0.19 |
ENST00000367688.3
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr7_+_150758642 | 0.19 |
ENST00000488420.1
|
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chrX_-_154688276 | 0.18 |
ENST00000369445.2
|
F8A3
|
coagulation factor VIII-associated 3 |
| chr9_-_140513231 | 0.18 |
ENST00000371417.3
|
C9orf37
|
chromosome 9 open reading frame 37 |
| chr17_-_46623441 | 0.18 |
ENST00000330070.4
|
HOXB2
|
homeobox B2 |
| chr2_-_72375167 | 0.18 |
ENST00000001146.2
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr1_+_16693578 | 0.18 |
ENST00000401088.4
ENST00000471507.1 ENST00000401089.3 ENST00000375590.3 ENST00000492354.1 |
SZRD1
|
SUZ RNA binding domain containing 1 |
| chr7_+_155250824 | 0.18 |
ENST00000297375.4
|
EN2
|
engrailed homeobox 2 |
| chr20_+_34680620 | 0.18 |
ENST00000430276.1
ENST00000373950.2 ENST00000452261.1 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr17_-_7297519 | 0.18 |
ENST00000576362.1
ENST00000571078.1 |
TMEM256-PLSCR3
|
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr12_-_6798025 | 0.18 |
ENST00000542351.1
ENST00000538829.1 |
ZNF384
|
zinc finger protein 384 |
| chr1_-_149908217 | 0.18 |
ENST00000369140.3
|
MTMR11
|
myotubularin related protein 11 |
| chr12_-_6798410 | 0.18 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr2_+_191045656 | 0.17 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr6_+_143771934 | 0.17 |
ENST00000367592.1
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr22_+_24891210 | 0.17 |
ENST00000382760.2
|
UPB1
|
ureidopropionase, beta |
| chr20_-_48532046 | 0.17 |
ENST00000543716.1
|
SPATA2
|
spermatogenesis associated 2 |
| chr12_-_6798616 | 0.17 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr10_+_72972281 | 0.17 |
ENST00000335350.6
|
UNC5B
|
unc-5 homolog B (C. elegans) |
| chr4_-_185139062 | 0.17 |
ENST00000296741.2
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr22_+_22676808 | 0.17 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr20_+_35974532 | 0.17 |
ENST00000373578.2
|
SRC
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr22_-_24641027 | 0.17 |
ENST00000398292.3
ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5
|
gamma-glutamyltransferase 5 |
| chr20_+_44098346 | 0.17 |
ENST00000372676.3
|
WFDC2
|
WAP four-disulfide core domain 2 |
| chr12_+_110338323 | 0.17 |
ENST00000312777.5
ENST00000536408.2 |
TCHP
|
trichoplein, keratin filament binding |
| chr20_+_42143053 | 0.16 |
ENST00000373135.3
ENST00000444063.1 |
L3MBTL1
|
l(3)mbt-like 1 (Drosophila) |
| chr16_+_53164956 | 0.16 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr4_-_103749179 | 0.16 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chrX_-_30595959 | 0.16 |
ENST00000378962.3
|
CXorf21
|
chromosome X open reading frame 21 |
| chr7_-_130080977 | 0.16 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr6_+_31730773 | 0.16 |
ENST00000415669.2
ENST00000425424.1 |
SAPCD1
|
suppressor APC domain containing 1 |
| chr3_-_49761337 | 0.16 |
ENST00000535833.1
ENST00000308388.6 ENST00000480687.1 ENST00000308375.6 |
AMIGO3
GMPPB
|
adhesion molecule with Ig-like domain 3 GDP-mannose pyrophosphorylase B |
| chrX_-_107018969 | 0.16 |
ENST00000372383.4
|
TSC22D3
|
TSC22 domain family, member 3 |
| chr11_+_74204395 | 0.16 |
ENST00000526036.1
|
AP001372.2
|
AP001372.2 |
| chr1_-_207119738 | 0.16 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr5_+_133861339 | 0.15 |
ENST00000282605.4
ENST00000361895.2 ENST00000402835.1 |
PHF15
|
jade family PHD finger 2 |
| chr19_-_42636617 | 0.15 |
ENST00000529067.1
ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2
|
POU class 2 homeobox 2 |
| chr20_-_48532019 | 0.15 |
ENST00000289431.5
|
SPATA2
|
spermatogenesis associated 2 |
| chr19_+_49128209 | 0.15 |
ENST00000599748.1
ENST00000443164.1 ENST00000599029.1 |
SPHK2
|
sphingosine kinase 2 |
| chr6_+_30853002 | 0.15 |
ENST00000421124.2
ENST00000512725.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr7_-_130080818 | 0.15 |
ENST00000343969.5
ENST00000541543.1 ENST00000489512.1 |
CEP41
|
centrosomal protein 41kDa |
| chr6_+_32938692 | 0.14 |
ENST00000443797.2
|
BRD2
|
bromodomain containing 2 |
| chr3_-_71179699 | 0.14 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr12_+_93965609 | 0.14 |
ENST00000549887.1
ENST00000551556.1 |
SOCS2
|
suppressor of cytokine signaling 2 |
| chr15_+_74509530 | 0.14 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr12_+_53443963 | 0.14 |
ENST00000546602.1
ENST00000552570.1 ENST00000549700.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr17_+_36508826 | 0.14 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr17_+_40761660 | 0.14 |
ENST00000251413.3
ENST00000591509.1 |
TUBG1
|
tubulin, gamma 1 |
| chr19_-_46288917 | 0.14 |
ENST00000537879.1
ENST00000596586.1 ENST00000595946.1 |
DMWD
AC011530.4
|
dystrophia myotonica, WD repeat containing Uncharacterized protein |
| chr18_+_3448455 | 0.13 |
ENST00000549780.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr9_-_136006496 | 0.13 |
ENST00000372062.3
|
RALGDS
|
ral guanine nucleotide dissociation stimulator |
| chr19_-_14606900 | 0.13 |
ENST00000393029.3
ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1
|
GIPC PDZ domain containing family, member 1 |
| chr5_-_64064508 | 0.13 |
ENST00000513458.4
|
SREK1IP1
|
SREK1-interacting protein 1 |
| chr19_+_56186606 | 0.13 |
ENST00000085079.7
|
EPN1
|
epsin 1 |
| chr1_-_94586651 | 0.13 |
ENST00000535735.1
ENST00000370225.3 |
ABCA4
|
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr12_-_120663792 | 0.12 |
ENST00000546532.1
ENST00000548912.1 |
PXN
|
paxillin |
| chr19_+_49977818 | 0.12 |
ENST00000594009.1
ENST00000595510.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr2_+_74881398 | 0.12 |
ENST00000339773.5
ENST00000434486.1 |
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr7_+_102004322 | 0.12 |
ENST00000496391.1
|
PRKRIP1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr12_-_71003568 | 0.12 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr5_+_139493665 | 0.12 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr6_+_4087664 | 0.11 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr11_-_75380165 | 0.11 |
ENST00000304771.3
|
MAP6
|
microtubule-associated protein 6 |
| chr3_-_49131013 | 0.11 |
ENST00000424300.1
|
QRICH1
|
glutamine-rich 1 |
| chr3_-_150320937 | 0.11 |
ENST00000479209.1
|
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr9_+_5890802 | 0.11 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr12_+_7037461 | 0.11 |
ENST00000396684.2
|
ATN1
|
atrophin 1 |
| chr19_-_46272462 | 0.11 |
ENST00000317578.6
|
SIX5
|
SIX homeobox 5 |
| chr9_-_28670283 | 0.11 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr12_-_120554534 | 0.10 |
ENST00000538903.1
ENST00000534951.1 |
RAB35
|
RAB35, member RAS oncogene family |
| chr21_-_38639813 | 0.10 |
ENST00000309117.6
ENST00000398998.1 |
DSCR3
|
Down syndrome critical region gene 3 |
| chr6_+_30689401 | 0.10 |
ENST00000396389.1
ENST00000396384.1 |
TUBB
|
tubulin, beta class I |
| chr17_-_1389419 | 0.10 |
ENST00000575158.1
|
MYO1C
|
myosin IC |
| chr9_+_34990219 | 0.10 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chrX_+_38420783 | 0.10 |
ENST00000422612.2
ENST00000286824.6 ENST00000545599.1 |
TSPAN7
|
tetraspanin 7 |
| chr17_+_40440094 | 0.10 |
ENST00000546010.2
|
STAT5A
|
signal transducer and activator of transcription 5A |
| chr16_+_2286726 | 0.10 |
ENST00000382437.4
ENST00000569184.1 |
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr20_+_30598231 | 0.10 |
ENST00000300415.8
ENST00000262659.8 |
CCM2L
|
cerebral cavernous malformation 2-like |
| chr16_+_30406423 | 0.10 |
ENST00000524644.1
|
ZNF48
|
zinc finger protein 48 |
| chr18_-_24237339 | 0.10 |
ENST00000580191.1
|
KCTD1
|
potassium channel tetramerization domain containing 1 |
| chr12_-_22094336 | 0.09 |
ENST00000326684.4
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr19_-_14224969 | 0.09 |
ENST00000589994.1
|
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr16_-_67997947 | 0.09 |
ENST00000537830.2
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr9_+_125273081 | 0.09 |
ENST00000335302.5
|
OR1J2
|
olfactory receptor, family 1, subfamily J, member 2 |
| chr16_+_2198604 | 0.09 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr11_-_34535332 | 0.09 |
ENST00000257832.2
ENST00000429939.2 |
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr10_-_14372870 | 0.09 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr2_-_74667612 | 0.09 |
ENST00000305557.5
ENST00000233330.6 |
RTKN
|
rhotekin |
| chr7_-_105517021 | 0.09 |
ENST00000318724.4
ENST00000419735.3 |
ATXN7L1
|
ataxin 7-like 1 |
| chr6_+_30852738 | 0.09 |
ENST00000508312.1
ENST00000512336.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_155271213 | 0.09 |
ENST00000342741.4
|
PKLR
|
pyruvate kinase, liver and RBC |
| chr8_-_142377367 | 0.09 |
ENST00000377741.3
|
GPR20
|
G protein-coupled receptor 20 |
| chr9_-_130829588 | 0.09 |
ENST00000373078.4
|
NAIF1
|
nuclear apoptosis inducing factor 1 |
| chr4_+_102268904 | 0.08 |
ENST00000527564.1
ENST00000529296.1 |
AP001816.1
|
Uncharacterized protein |
| chr16_-_56459354 | 0.08 |
ENST00000290649.5
|
AMFR
|
autocrine motility factor receptor, E3 ubiquitin protein ligase |
| chr9_+_133569108 | 0.08 |
ENST00000372358.5
ENST00000546165.1 ENST00000372352.3 ENST00000372351.3 ENST00000372350.3 ENST00000495699.2 |
EXOSC2
|
exosome component 2 |
| chr1_+_26872324 | 0.08 |
ENST00000531382.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr17_+_3627185 | 0.08 |
ENST00000325418.4
|
GSG2
|
germ cell associated 2 (haspin) |
| chr2_+_191045562 | 0.08 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr17_-_10017864 | 0.08 |
ENST00000323816.4
|
GAS7
|
growth arrest-specific 7 |
| chr11_-_74204742 | 0.07 |
ENST00000310109.4
|
LIPT2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr4_+_39640787 | 0.07 |
ENST00000532680.1
|
RP11-539G18.2
|
RP11-539G18.2 |
| chr9_+_71944241 | 0.07 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr12_+_68042517 | 0.07 |
ENST00000393555.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr16_+_58059470 | 0.07 |
ENST00000219271.3
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr12_+_53443680 | 0.07 |
ENST00000314250.6
ENST00000451358.1 |
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2) |
| chr2_+_27237615 | 0.07 |
ENST00000458529.1
ENST00000402218.1 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr5_+_175298573 | 0.07 |
ENST00000512824.1
|
CPLX2
|
complexin 2 |
| chr6_-_31612808 | 0.07 |
ENST00000438149.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr4_-_41216473 | 0.07 |
ENST00000513140.1
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr4_-_102268708 | 0.07 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr18_+_13217965 | 0.06 |
ENST00000587905.1
|
LDLRAD4
|
low density lipoprotein receptor class A domain containing 4 |
| chrX_-_107019181 | 0.06 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr1_+_100111479 | 0.06 |
ENST00000263174.4
|
PALMD
|
palmdelphin |
| chrX_+_153029633 | 0.06 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr11_-_1619524 | 0.06 |
ENST00000412090.1
|
KRTAP5-2
|
keratin associated protein 5-2 |
| chr15_-_88799661 | 0.06 |
ENST00000360948.2
ENST00000357724.2 ENST00000355254.2 ENST00000317501.3 |
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr16_-_17564738 | 0.06 |
ENST00000261381.6
|
XYLT1
|
xylosyltransferase I |
| chrY_+_16634483 | 0.06 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr22_-_31885514 | 0.06 |
ENST00000397525.1
|
EIF4ENIF1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr17_+_7792101 | 0.05 |
ENST00000358181.4
ENST00000330494.7 |
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr1_+_156163880 | 0.05 |
ENST00000359511.4
ENST00000423538.2 |
SLC25A44
|
solute carrier family 25, member 44 |
| chr1_+_15802594 | 0.05 |
ENST00000375910.3
|
CELA2B
|
chymotrypsin-like elastase family, member 2B |
| chrX_+_591524 | 0.05 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr17_-_15165854 | 0.05 |
ENST00000395936.1
ENST00000395938.2 |
PMP22
|
peripheral myelin protein 22 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SMAD2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0060738 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of immature T cell proliferation in thymus(GO:0033092) right lung development(GO:0060458) primary prostatic bud elongation(GO:0060516) epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.2 | 0.6 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.2 | 0.5 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.2 | 0.9 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.4 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.2 | GO:2001037 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 0.2 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.3 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.2 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.7 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.6 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.3 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0048691 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) regulation of osteoclast proliferation(GO:0090289) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.3 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:1904616 | regulation of actin filament binding(GO:1904529) negative regulation of actin filament binding(GO:1904530) regulation of actin binding(GO:1904616) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.3 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070931 | Golgi-associated vesicle lumen(GO:0070931) |
| 0.1 | 0.6 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.9 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.7 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 1.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.4 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 0.2 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.1 | 0.4 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.2 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.0 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 1.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |