Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
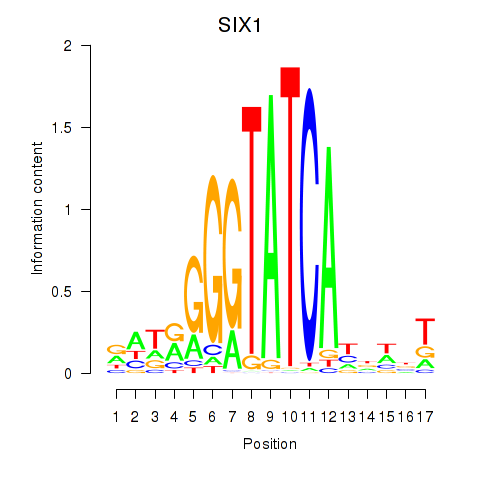
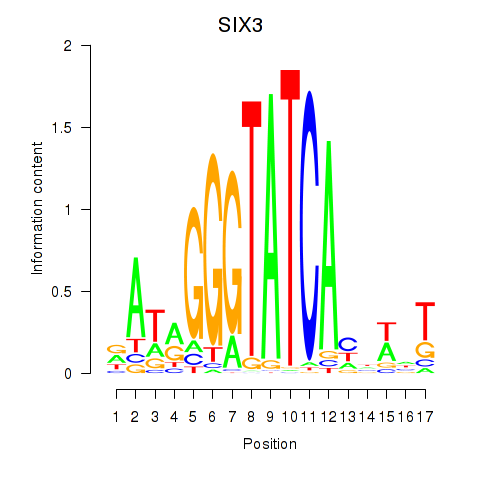
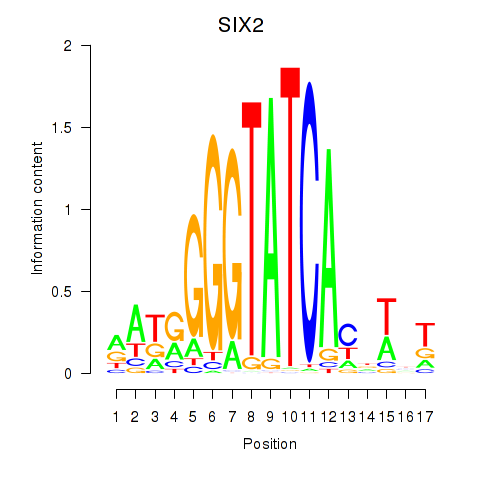
Results for SIX1_SIX3_SIX2
Z-value: 0.83
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.7 | SIX homeobox 1 |
|
SIX3
|
ENSG00000138083.3 | SIX homeobox 3 |
|
SIX2
|
ENSG00000170577.7 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX1 | hg19_v2_chr14_-_61124977_61125037 | 0.86 | 3.0e-02 | Click! |
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | 0.41 | 4.2e-01 | Click! |
| SIX2 | hg19_v2_chr2_-_45236540_45236577 | 0.06 | 9.1e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_28550320 | 0.89 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr3_+_42850959 | 0.72 |
ENST00000442925.1
ENST00000422265.1 ENST00000497921.1 ENST00000426937.1 |
ACKR2
KRBOX1
|
atypical chemokine receptor 2 KRAB box domain containing 1 |
| chr7_+_139025875 | 0.69 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr14_+_91709103 | 0.66 |
ENST00000553725.1
|
CTD-2547L24.3
|
HCG1816139; Uncharacterized protein |
| chr1_-_179457805 | 0.51 |
ENST00000600581.1
|
AL160286.1
|
Uncharacterized protein |
| chr16_-_28550348 | 0.49 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr3_+_70048881 | 0.46 |
ENST00000483525.1
|
RP11-460N16.1
|
RP11-460N16.1 |
| chr13_-_46679144 | 0.39 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr7_-_34978980 | 0.32 |
ENST00000428054.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr16_+_58010339 | 0.32 |
ENST00000290871.5
ENST00000441824.2 |
TEPP
|
testis, prostate and placenta expressed |
| chr10_-_33405600 | 0.32 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chr1_-_44818599 | 0.31 |
ENST00000537474.1
|
ERI3
|
ERI1 exoribonuclease family member 3 |
| chr6_+_26273144 | 0.31 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr7_-_48068699 | 0.29 |
ENST00000412142.1
ENST00000395572.2 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr1_+_241695670 | 0.29 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_-_50990785 | 0.28 |
ENST00000595005.1
|
CTD-2545M3.8
|
CTD-2545M3.8 |
| chr20_+_34824355 | 0.28 |
ENST00000397286.3
ENST00000320849.4 ENST00000373932.3 |
AAR2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr12_-_58220078 | 0.28 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr14_-_24911448 | 0.27 |
ENST00000555355.1
ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr14_+_75761099 | 0.27 |
ENST00000561000.1
ENST00000558575.1 |
RP11-293M10.5
|
RP11-293M10.5 |
| chr5_+_20616500 | 0.26 |
ENST00000512688.1
|
RP11-774D14.1
|
RP11-774D14.1 |
| chr1_+_150245177 | 0.25 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr22_+_19710468 | 0.25 |
ENST00000366425.3
|
GP1BB
|
glycoprotein Ib (platelet), beta polypeptide |
| chr1_+_241695424 | 0.24 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr15_-_31521567 | 0.22 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr4_+_2794750 | 0.22 |
ENST00000452765.2
ENST00000389838.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr1_-_158656488 | 0.22 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr1_+_156698743 | 0.21 |
ENST00000524343.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr20_+_62694834 | 0.21 |
ENST00000415602.1
|
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr14_-_95942173 | 0.21 |
ENST00000334258.5
ENST00000557275.1 ENST00000553340.1 |
SYNE3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr16_+_29690358 | 0.21 |
ENST00000395384.4
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr16_+_19098178 | 0.20 |
ENST00000568032.1
|
RP11-626G11.4
|
RP11-626G11.4 |
| chr17_-_55911970 | 0.20 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr2_-_100925967 | 0.20 |
ENST00000409647.1
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2 |
| chr13_-_30160925 | 0.20 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr14_-_74296806 | 0.20 |
ENST00000555539.1
|
RP5-1021I20.2
|
RP5-1021I20.2 |
| chr20_-_4055812 | 0.19 |
ENST00000379526.1
|
RP11-352D3.2
|
Uncharacterized protein |
| chr3_+_184530173 | 0.19 |
ENST00000453056.1
|
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr11_-_104769141 | 0.19 |
ENST00000508062.1
ENST00000422698.2 |
CASP12
|
caspase 12 (gene/pseudogene) |
| chr6_+_116782527 | 0.18 |
ENST00000368606.3
ENST00000368605.1 |
FAM26F
|
family with sequence similarity 26, member F |
| chr17_-_33446735 | 0.18 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr5_+_125759140 | 0.18 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_79783788 | 0.18 |
ENST00000282226.4
|
FAM151B
|
family with sequence similarity 151, member B |
| chr19_-_11039261 | 0.18 |
ENST00000590329.1
ENST00000587943.1 ENST00000585858.1 ENST00000586748.1 ENST00000586575.1 ENST00000253031.2 |
YIPF2
|
Yip1 domain family, member 2 |
| chr16_+_85936295 | 0.18 |
ENST00000563180.1
ENST00000564617.1 ENST00000564803.1 |
IRF8
|
interferon regulatory factor 8 |
| chr16_-_69448 | 0.17 |
ENST00000326592.9
|
WASH4P
|
WAS protein family homolog 4 pseudogene |
| chr12_-_118796910 | 0.17 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr12_+_110011571 | 0.17 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr1_+_158901329 | 0.17 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr7_-_48068671 | 0.17 |
ENST00000297325.4
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr3_+_129207033 | 0.17 |
ENST00000507221.1
|
IFT122
|
intraflagellar transport 122 homolog (Chlamydomonas) |
| chr5_+_154173697 | 0.17 |
ENST00000518742.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr16_+_32264645 | 0.17 |
ENST00000569631.1
ENST00000354614.3 |
TP53TG3D
|
TP53 target 3D |
| chr12_-_53901266 | 0.17 |
ENST00000609999.1
ENST00000267017.3 |
NPFF
|
neuropeptide FF-amide peptide precursor |
| chr15_+_62853562 | 0.16 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr10_-_49860525 | 0.16 |
ENST00000435790.2
|
ARHGAP22
|
Rho GTPase activating protein 22 |
| chr4_-_140477928 | 0.16 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr16_+_22517166 | 0.16 |
ENST00000356156.3
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr1_-_54665664 | 0.16 |
ENST00000542737.1
ENST00000537208.1 |
CYB5RL
|
cytochrome b5 reductase-like |
| chr17_+_4675175 | 0.16 |
ENST00000270560.3
|
TM4SF5
|
transmembrane 4 L six family member 5 |
| chr17_-_7080883 | 0.15 |
ENST00000570576.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr11_+_82783097 | 0.15 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr3_-_157251383 | 0.15 |
ENST00000487753.1
ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr12_+_57624059 | 0.15 |
ENST00000557427.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr19_+_47523058 | 0.15 |
ENST00000602212.1
ENST00000602189.1 |
NPAS1
|
neuronal PAS domain protein 1 |
| chr11_+_70244510 | 0.15 |
ENST00000346329.3
ENST00000301843.8 ENST00000376561.3 |
CTTN
|
cortactin |
| chr4_+_11470867 | 0.15 |
ENST00000515343.1
|
RP11-281P23.1
|
RP11-281P23.1 |
| chr7_+_100551239 | 0.15 |
ENST00000319509.7
|
MUC3A
|
mucin 3A, cell surface associated |
| chr10_+_135207598 | 0.14 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr17_+_79031415 | 0.14 |
ENST00000572073.1
ENST00000573677.1 |
BAIAP2
|
BAI1-associated protein 2 |
| chr19_+_11457232 | 0.14 |
ENST00000587531.1
|
CCDC159
|
coiled-coil domain containing 159 |
| chr1_+_160709055 | 0.14 |
ENST00000368043.3
ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7
|
SLAM family member 7 |
| chr19_+_36632056 | 0.14 |
ENST00000586851.1
ENST00000590211.1 |
CAPNS1
|
calpain, small subunit 1 |
| chr21_+_27011584 | 0.14 |
ENST00000400532.1
ENST00000480456.1 ENST00000312957.5 |
JAM2
|
junctional adhesion molecule 2 |
| chr9_+_130830451 | 0.14 |
ENST00000373068.2
ENST00000373069.5 |
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr6_+_31802364 | 0.14 |
ENST00000375640.3
ENST00000375641.2 |
C6orf48
|
chromosome 6 open reading frame 48 |
| chr1_+_54703710 | 0.13 |
ENST00000361350.4
|
SSBP3-AS1
|
SSBP3 antisense RNA 1 |
| chr3_-_11685345 | 0.13 |
ENST00000430365.2
|
VGLL4
|
vestigial like 4 (Drosophila) |
| chr22_-_32767017 | 0.13 |
ENST00000400234.1
|
RFPL3S
|
RFPL3 antisense |
| chr19_+_42055879 | 0.13 |
ENST00000407170.2
ENST00000601116.1 ENST00000595395.1 |
CEACAM21
AC006129.2
|
carcinoembryonic antigen-related cell adhesion molecule 21 AC006129.2 |
| chr14_-_24729251 | 0.13 |
ENST00000559136.1
|
TGM1
|
transglutaminase 1 |
| chr12_+_57623907 | 0.13 |
ENST00000553529.1
ENST00000554310.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr15_+_40650408 | 0.13 |
ENST00000267889.3
|
DISP2
|
dispatched homolog 2 (Drosophila) |
| chr7_-_14880892 | 0.13 |
ENST00000406247.3
ENST00000399322.3 ENST00000258767.5 |
DGKB
|
diacylglycerol kinase, beta 90kDa |
| chr16_-_2581409 | 0.13 |
ENST00000567119.1
ENST00000565480.1 ENST00000382350.1 |
CEMP1
|
cementum protein 1 |
| chr11_-_2906979 | 0.13 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr16_+_68279256 | 0.13 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr12_-_112614506 | 0.13 |
ENST00000548588.2
|
HECTD4
|
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr15_+_75491213 | 0.13 |
ENST00000360639.2
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr12_-_11036844 | 0.12 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr10_+_48359344 | 0.12 |
ENST00000412534.1
ENST00000444585.1 |
ZNF488
|
zinc finger protein 488 |
| chr19_+_36631867 | 0.12 |
ENST00000588780.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr2_-_220034745 | 0.12 |
ENST00000455516.2
ENST00000396775.3 ENST00000295738.7 |
SLC23A3
|
solute carrier family 23, member 3 |
| chr11_-_59633951 | 0.12 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr21_+_38792602 | 0.12 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr7_+_149570049 | 0.12 |
ENST00000421974.2
ENST00000456496.2 |
ATP6V0E2
|
ATPase, H+ transporting V0 subunit e2 |
| chr15_+_40331456 | 0.12 |
ENST00000504245.1
ENST00000560341.1 |
SRP14-AS1
|
SRP14 antisense RNA1 (head to head) |
| chr3_-_9595480 | 0.12 |
ENST00000287585.6
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4 |
| chr5_+_125758813 | 0.12 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr11_-_130786333 | 0.12 |
ENST00000533214.1
ENST00000528555.1 ENST00000530356.1 ENST00000539184.1 |
SNX19
|
sorting nexin 19 |
| chr8_-_83589388 | 0.11 |
ENST00000522776.1
|
RP11-653B10.1
|
RP11-653B10.1 |
| chr12_-_9913489 | 0.11 |
ENST00000228434.3
ENST00000536709.1 |
CD69
|
CD69 molecule |
| chr16_+_57673207 | 0.11 |
ENST00000564783.1
ENST00000564729.1 ENST00000565976.1 ENST00000566508.1 ENST00000544297.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr7_+_150759634 | 0.11 |
ENST00000392826.2
ENST00000461735.1 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr10_+_135207623 | 0.11 |
ENST00000317502.6
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr6_-_32160622 | 0.11 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chr4_+_89514516 | 0.11 |
ENST00000452979.1
|
HERC3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr19_+_14551066 | 0.11 |
ENST00000342216.4
|
PKN1
|
protein kinase N1 |
| chr12_+_57624085 | 0.11 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr16_+_27214802 | 0.11 |
ENST00000380948.2
ENST00000286096.4 |
KDM8
|
lysine (K)-specific demethylase 8 |
| chr6_-_167797887 | 0.11 |
ENST00000476779.2
ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10
|
t-complex 10 |
| chr4_+_89300158 | 0.11 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr1_-_150979333 | 0.11 |
ENST00000312210.5
|
FAM63A
|
family with sequence similarity 63, member A |
| chr1_+_12916941 | 0.11 |
ENST00000240189.2
|
PRAMEF2
|
PRAME family member 2 |
| chr11_-_8892900 | 0.11 |
ENST00000526155.1
ENST00000524757.1 ENST00000527392.1 ENST00000534665.1 ENST00000525169.1 ENST00000527516.1 ENST00000533471.1 |
ST5
|
suppression of tumorigenicity 5 |
| chr12_-_111395610 | 0.11 |
ENST00000548329.1
ENST00000546852.1 |
RP1-46F2.3
|
RP1-46F2.3 |
| chr19_-_3600549 | 0.11 |
ENST00000589966.1
|
TBXA2R
|
thromboxane A2 receptor |
| chr19_+_50270219 | 0.11 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr5_+_125758865 | 0.11 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr16_+_28763108 | 0.10 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr11_-_71791518 | 0.10 |
ENST00000537217.1
ENST00000366394.3 ENST00000358965.6 ENST00000546131.1 ENST00000543937.1 ENST00000368959.5 ENST00000541641.1 |
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr12_+_90674665 | 0.10 |
ENST00000549313.1
|
RP11-753N8.1
|
RP11-753N8.1 |
| chr8_+_19536083 | 0.10 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr9_+_12693336 | 0.10 |
ENST00000381137.2
ENST00000388918.5 |
TYRP1
|
tyrosinase-related protein 1 |
| chr19_+_36605850 | 0.10 |
ENST00000221855.3
|
TBCB
|
tubulin folding cofactor B |
| chr7_+_151771377 | 0.10 |
ENST00000434507.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr19_+_11039391 | 0.10 |
ENST00000270502.6
|
C19orf52
|
chromosome 19 open reading frame 52 |
| chr19_+_7981030 | 0.10 |
ENST00000565886.1
|
TGFBR3L
|
transforming growth factor, beta receptor III-like |
| chr12_+_57624119 | 0.10 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr5_-_141392538 | 0.10 |
ENST00000503794.1
ENST00000510194.1 ENST00000504424.1 ENST00000513454.1 ENST00000458112.2 ENST00000542860.1 ENST00000503229.1 ENST00000500692.2 ENST00000311337.6 ENST00000504139.1 ENST00000505689.1 |
GNPDA1
|
glucosamine-6-phosphate deaminase 1 |
| chr1_-_51791596 | 0.10 |
ENST00000532836.1
ENST00000422925.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr1_+_150245099 | 0.10 |
ENST00000369099.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr15_+_80733570 | 0.10 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr1_+_149239529 | 0.10 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr11_-_34533257 | 0.10 |
ENST00000312319.2
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr21_-_33957805 | 0.10 |
ENST00000300258.3
ENST00000472557.1 |
TCP10L
|
t-complex 10-like |
| chr5_+_140705777 | 0.10 |
ENST00000606901.1
ENST00000606674.1 |
AC005618.6
|
AC005618.6 |
| chr19_+_17420340 | 0.10 |
ENST00000359866.4
|
DDA1
|
DET1 and DDB1 associated 1 |
| chr17_-_41466555 | 0.10 |
ENST00000586231.1
|
LINC00910
|
long intergenic non-protein coding RNA 910 |
| chr5_+_81575281 | 0.10 |
ENST00000380167.4
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr10_-_46030841 | 0.09 |
ENST00000453424.2
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr2_+_74361599 | 0.09 |
ENST00000401851.1
|
MGC10955
|
MGC10955 |
| chr1_+_156698708 | 0.09 |
ENST00000519086.1
|
RRNAD1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr12_+_57623869 | 0.09 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr6_+_160693591 | 0.09 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr4_-_52883786 | 0.09 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr7_+_141490017 | 0.09 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr7_+_72349920 | 0.09 |
ENST00000395270.1
ENST00000446813.1 ENST00000257622.4 |
POM121
|
POM121 transmembrane nucleoporin |
| chr19_+_56116771 | 0.09 |
ENST00000568956.1
|
ZNF865
|
zinc finger protein 865 |
| chr1_-_207119738 | 0.09 |
ENST00000356495.4
|
PIGR
|
polymeric immunoglobulin receptor |
| chr11_+_125365110 | 0.09 |
ENST00000527818.1
|
AP000708.1
|
AP000708.1 |
| chr11_+_63742050 | 0.09 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr3_-_178984759 | 0.09 |
ENST00000349697.2
ENST00000497599.1 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr1_+_6684918 | 0.09 |
ENST00000054650.4
|
THAP3
|
THAP domain containing, apoptosis associated protein 3 |
| chr4_-_860950 | 0.08 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr16_-_67867749 | 0.08 |
ENST00000566758.1
ENST00000445712.2 ENST00000219172.3 ENST00000564817.1 |
CENPT
|
centromere protein T |
| chr12_+_7053228 | 0.08 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr8_-_123139423 | 0.08 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr12_-_50790267 | 0.08 |
ENST00000327337.5
ENST00000543111.1 |
FAM186A
|
family with sequence similarity 186, member A |
| chr9_-_95186739 | 0.08 |
ENST00000375550.4
|
OMD
|
osteomodulin |
| chr19_-_37407172 | 0.08 |
ENST00000391711.3
|
ZNF829
|
zinc finger protein 829 |
| chr12_+_49717019 | 0.08 |
ENST00000549275.1
ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP
|
trophinin associated protein |
| chr19_+_36606354 | 0.08 |
ENST00000589996.1
ENST00000591296.1 |
TBCB
|
tubulin folding cofactor B |
| chr17_-_8113542 | 0.08 |
ENST00000578549.1
ENST00000535053.1 ENST00000582368.1 |
AURKB
|
aurora kinase B |
| chr19_-_36606181 | 0.08 |
ENST00000221859.4
|
POLR2I
|
polymerase (RNA) II (DNA directed) polypeptide I, 14.5kDa |
| chr20_-_33732952 | 0.08 |
ENST00000541621.1
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr12_-_10826612 | 0.08 |
ENST00000535345.1
ENST00000542562.1 |
STYK1
|
serine/threonine/tyrosine kinase 1 |
| chr11_+_36317830 | 0.08 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr17_+_75315654 | 0.08 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr3_-_149095652 | 0.08 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chr12_+_69186125 | 0.08 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr2_-_3595547 | 0.08 |
ENST00000438485.1
|
RP13-512J5.1
|
Uncharacterized protein |
| chr17_+_7210294 | 0.08 |
ENST00000336452.7
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr15_+_89631647 | 0.08 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr1_+_53793885 | 0.08 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr14_-_24740709 | 0.08 |
ENST00000399409.3
ENST00000216840.6 |
RABGGTA
|
Rab geranylgeranyltransferase, alpha subunit |
| chr19_+_45445524 | 0.08 |
ENST00000591600.1
|
APOC4
|
apolipoprotein C-IV |
| chr19_+_36632204 | 0.08 |
ENST00000592354.1
|
CAPNS1
|
calpain, small subunit 1 |
| chr17_+_1665306 | 0.08 |
ENST00000571360.1
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr19_+_17579556 | 0.07 |
ENST00000442725.1
|
SLC27A1
|
solute carrier family 27 (fatty acid transporter), member 1 |
| chr12_+_7053172 | 0.07 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr9_-_27005686 | 0.07 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr19_-_10946949 | 0.07 |
ENST00000214869.2
ENST00000591695.1 |
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr16_-_5115913 | 0.07 |
ENST00000474471.3
|
C16orf89
|
chromosome 16 open reading frame 89 |
| chr19_-_11039188 | 0.07 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr12_+_49717081 | 0.07 |
ENST00000547807.1
ENST00000551567.1 |
TROAP
|
trophinin associated protein |
| chr11_-_58378759 | 0.07 |
ENST00000601906.1
|
AP001350.1
|
Uncharacterized protein |
| chr19_+_49109990 | 0.07 |
ENST00000321762.1
|
SPACA4
|
sperm acrosome associated 4 |
| chr1_+_196743912 | 0.07 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr6_-_31509714 | 0.07 |
ENST00000456662.1
ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr6_-_31671058 | 0.07 |
ENST00000538874.1
ENST00000395952.3 |
ABHD16A
|
abhydrolase domain containing 16A |
| chr9_+_131549483 | 0.07 |
ENST00000372648.5
ENST00000539497.1 |
TBC1D13
|
TBC1 domain family, member 13 |
| chr17_-_4938712 | 0.07 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr16_+_84801852 | 0.07 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr12_-_6798616 | 0.07 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr9_+_131549610 | 0.07 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr17_-_6915646 | 0.07 |
ENST00000574377.1
ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2
|
Uncharacterized protein |
| chr1_+_174670143 | 0.07 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr6_-_31509506 | 0.07 |
ENST00000449757.1
|
DDX39B
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr11_+_71900572 | 0.07 |
ENST00000312293.4
|
FOLR1
|
folate receptor 1 (adult) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 0.7 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.1 | 1.4 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.6 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.2 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.2 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.0 | 0.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.3 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.2 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.2 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.0 | 0.1 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:0035934 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.0 | 0.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:2000570 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.1 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.1 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:1902363 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.0 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.0 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0036026 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.0 | 0.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.6 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |