Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
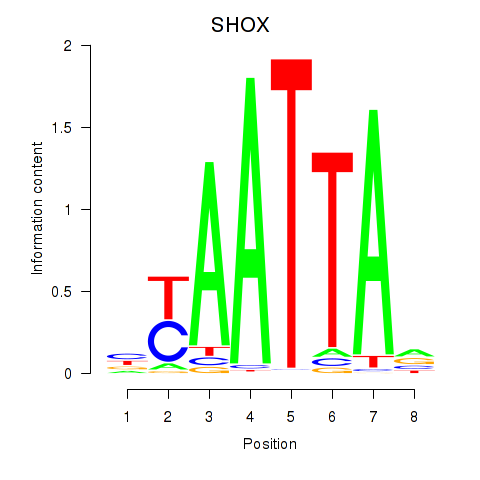
Results for SHOX
Z-value: 0.49
Transcription factors associated with SHOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SHOX
|
ENSG00000185960.8 | short stature homeobox |
|
SHOX
|
ENSGR0000185960.8 | short stature homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SHOX | hg19_v2_chrX_+_591524_591542 | 0.49 | 3.2e-01 | Click! |
Activity profile of SHOX motif
Sorted Z-values of SHOX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_89746264 | 0.32 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr17_+_19091325 | 0.31 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr17_-_38821373 | 0.31 |
ENST00000394052.3
|
KRT222
|
keratin 222 |
| chr4_+_183065793 | 0.28 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr14_-_57197224 | 0.25 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr14_+_20187174 | 0.25 |
ENST00000557414.1
|
OR4N2
|
olfactory receptor, family 4, subfamily N, member 2 |
| chr5_-_24645078 | 0.24 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr4_+_95916947 | 0.24 |
ENST00000506363.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr7_-_92777606 | 0.23 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr12_-_16761117 | 0.23 |
ENST00000538051.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr14_-_98444386 | 0.22 |
ENST00000556462.1
ENST00000556138.1 |
C14orf64
|
chromosome 14 open reading frame 64 |
| chr11_+_77532233 | 0.22 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr17_-_39341594 | 0.21 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr3_-_27763803 | 0.21 |
ENST00000449599.1
|
EOMES
|
eomesodermin |
| chr6_-_31107127 | 0.20 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chrM_+_10464 | 0.19 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr12_+_116955659 | 0.19 |
ENST00000552992.1
|
RP11-148B3.1
|
RP11-148B3.1 |
| chr2_+_172309634 | 0.19 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr4_-_36245561 | 0.18 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr13_-_88323218 | 0.18 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr12_-_10022735 | 0.18 |
ENST00000228438.2
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr5_+_136070614 | 0.18 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chrX_+_68835911 | 0.18 |
ENST00000525810.1
ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA
|
ectodysplasin A |
| chrM_+_10758 | 0.18 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr16_+_53133070 | 0.17 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr20_-_50418947 | 0.17 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr3_-_27764190 | 0.16 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr5_+_174151536 | 0.16 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr17_-_19015945 | 0.16 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr17_-_40337470 | 0.16 |
ENST00000293330.1
|
HCRT
|
hypocretin (orexin) neuropeptide precursor |
| chr7_-_14026063 | 0.15 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr5_-_126409159 | 0.15 |
ENST00000607731.1
ENST00000535381.1 ENST00000296662.5 ENST00000509733.3 |
C5orf63
|
chromosome 5 open reading frame 63 |
| chr9_+_82188077 | 0.14 |
ENST00000425506.1
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_+_225600404 | 0.14 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr8_+_119294456 | 0.14 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr7_-_92747269 | 0.14 |
ENST00000446617.1
ENST00000379958.2 |
SAMD9
|
sterile alpha motif domain containing 9 |
| chrX_+_591524 | 0.14 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr9_-_3469181 | 0.13 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr17_+_56833184 | 0.13 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr16_+_53412368 | 0.13 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr12_-_88974236 | 0.13 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr1_+_186265399 | 0.12 |
ENST00000367486.3
ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4
|
proteoglycan 4 |
| chr5_+_31193847 | 0.12 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr8_+_94752349 | 0.12 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr10_-_21186144 | 0.12 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr6_+_105404899 | 0.12 |
ENST00000345080.4
|
LIN28B
|
lin-28 homolog B (C. elegans) |
| chr4_-_105416039 | 0.11 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr1_+_62439037 | 0.11 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr8_+_42873548 | 0.11 |
ENST00000533338.1
ENST00000534420.1 |
HOOK3
RP11-598P20.5
|
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr8_-_109799793 | 0.11 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr17_+_41363854 | 0.11 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr7_-_37488547 | 0.11 |
ENST00000453399.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr3_-_62359180 | 0.11 |
ENST00000283268.3
|
FEZF2
|
FEZ family zinc finger 2 |
| chr8_-_42358742 | 0.11 |
ENST00000517366.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr21_+_17443434 | 0.11 |
ENST00000400178.2
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chrM_+_7586 | 0.10 |
ENST00000361739.1
|
MT-CO2
|
mitochondrially encoded cytochrome c oxidase II |
| chr2_+_234826016 | 0.10 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr8_+_38831683 | 0.10 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr12_-_10282742 | 0.10 |
ENST00000298523.5
ENST00000396484.2 ENST00000310002.4 |
CLEC7A
|
C-type lectin domain family 7, member A |
| chr4_-_111563279 | 0.10 |
ENST00000511837.1
|
PITX2
|
paired-like homeodomain 2 |
| chr3_-_12587055 | 0.10 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr12_+_1099675 | 0.10 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr14_-_38036271 | 0.10 |
ENST00000556024.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr11_+_35222629 | 0.10 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr11_+_107461804 | 0.10 |
ENST00000531234.1
|
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr12_-_74686314 | 0.10 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr3_+_157154578 | 0.10 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr4_+_86748898 | 0.09 |
ENST00000509300.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr12_-_86650077 | 0.09 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr13_-_86373536 | 0.09 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr4_+_86749045 | 0.09 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr2_-_61697862 | 0.09 |
ENST00000398571.2
|
USP34
|
ubiquitin specific peptidase 34 |
| chr12_-_28122980 | 0.09 |
ENST00000395868.3
ENST00000534890.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr1_-_168464875 | 0.09 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr2_-_44550441 | 0.08 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr17_-_71223839 | 0.08 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr11_+_112041253 | 0.08 |
ENST00000532612.1
|
AP002884.3
|
AP002884.3 |
| chr15_-_55562479 | 0.08 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr12_-_89746173 | 0.08 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr16_-_58004992 | 0.08 |
ENST00000564448.1
ENST00000251102.8 ENST00000311183.4 |
CNGB1
|
cyclic nucleotide gated channel beta 1 |
| chr14_-_38028689 | 0.08 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr12_-_28123206 | 0.08 |
ENST00000542963.1
ENST00000535992.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr7_-_99716914 | 0.08 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr12_-_16759440 | 0.08 |
ENST00000537304.1
|
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr14_+_57671888 | 0.08 |
ENST00000391612.1
|
AL391152.1
|
AL391152.1 |
| chr1_-_68698197 | 0.08 |
ENST00000370973.2
ENST00000370971.1 |
WLS
|
wntless Wnt ligand secretion mediator |
| chr12_+_78359999 | 0.08 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr4_-_109541610 | 0.08 |
ENST00000510212.1
|
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr14_+_72052983 | 0.07 |
ENST00000358550.2
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr11_-_117747434 | 0.07 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr12_-_21928515 | 0.07 |
ENST00000537950.1
|
KCNJ8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr12_+_14561422 | 0.07 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr7_-_87342564 | 0.07 |
ENST00000265724.3
ENST00000416177.1 |
ABCB1
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr1_+_40713573 | 0.07 |
ENST00000372766.3
|
TMCO2
|
transmembrane and coiled-coil domains 2 |
| chr12_-_102591604 | 0.07 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr11_+_118398178 | 0.07 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr7_-_111424462 | 0.07 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr13_-_99910620 | 0.07 |
ENST00000416594.1
|
GPR18
|
G protein-coupled receptor 18 |
| chr2_+_190541153 | 0.07 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr7_-_14026123 | 0.07 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr10_-_104866395 | 0.07 |
ENST00000458345.1
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr8_-_116681221 | 0.07 |
ENST00000395715.3
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr1_-_101360331 | 0.07 |
ENST00000416479.1
ENST00000370113.3 |
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr2_-_145278475 | 0.07 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr8_-_33457453 | 0.07 |
ENST00000523956.1
ENST00000256261.4 |
DUSP26
|
dual specificity phosphatase 26 (putative) |
| chrM_+_3299 | 0.07 |
ENST00000361390.2
|
MT-ND1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr3_-_33686925 | 0.07 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr15_+_63188009 | 0.06 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr1_+_12042015 | 0.06 |
ENST00000412236.1
|
MFN2
|
mitofusin 2 |
| chr17_-_38928414 | 0.06 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chr2_-_203735484 | 0.06 |
ENST00000420558.1
ENST00000418208.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr17_-_62499334 | 0.06 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr3_+_141106458 | 0.06 |
ENST00000509883.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr9_-_16728161 | 0.06 |
ENST00000603713.1
ENST00000603313.1 |
BNC2
|
basonuclin 2 |
| chr12_+_52695617 | 0.06 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr3_+_141105705 | 0.06 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr1_+_84767289 | 0.06 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr6_+_5261225 | 0.06 |
ENST00000324331.6
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr1_-_151762900 | 0.06 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr7_-_14029515 | 0.06 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr6_-_39693111 | 0.06 |
ENST00000373215.3
ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6
|
kinesin family member 6 |
| chr15_-_55563072 | 0.06 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr11_-_33913708 | 0.05 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr15_-_55562451 | 0.05 |
ENST00000568803.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr13_+_78109884 | 0.05 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chrX_+_77166172 | 0.05 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr15_-_55562582 | 0.05 |
ENST00000396307.2
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr2_-_145277569 | 0.05 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr19_-_7968427 | 0.05 |
ENST00000539278.1
|
AC010336.1
|
Uncharacterized protein |
| chr4_-_170897045 | 0.05 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chrX_-_71458802 | 0.05 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr7_-_28220354 | 0.05 |
ENST00000283928.5
|
JAZF1
|
JAZF zinc finger 1 |
| chr9_+_124329336 | 0.05 |
ENST00000394340.3
ENST00000436835.1 ENST00000259371.2 |
DAB2IP
|
DAB2 interacting protein |
| chr18_-_53253112 | 0.05 |
ENST00000568673.1
ENST00000562847.1 ENST00000568147.1 |
TCF4
|
transcription factor 4 |
| chr4_-_69111401 | 0.05 |
ENST00000332644.5
|
TMPRSS11B
|
transmembrane protease, serine 11B |
| chr12_+_54378923 | 0.05 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr12_+_81110684 | 0.05 |
ENST00000228644.3
|
MYF5
|
myogenic factor 5 |
| chr17_-_38956205 | 0.05 |
ENST00000306658.7
|
KRT28
|
keratin 28 |
| chr5_-_20575959 | 0.05 |
ENST00000507958.1
|
CDH18
|
cadherin 18, type 2 |
| chr15_-_33180439 | 0.05 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr18_-_53253323 | 0.05 |
ENST00000540999.1
ENST00000563888.2 |
TCF4
|
transcription factor 4 |
| chrX_+_43515467 | 0.05 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr2_+_170440844 | 0.05 |
ENST00000260970.3
ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG
|
peptidylprolyl isomerase G (cyclophilin G) |
| chr8_+_117950422 | 0.05 |
ENST00000378279.3
|
AARD
|
alanine and arginine rich domain containing protein |
| chr4_+_90033968 | 0.05 |
ENST00000317005.2
|
TIGD2
|
tigger transposable element derived 2 |
| chr1_+_174844645 | 0.05 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr14_-_92413353 | 0.05 |
ENST00000556154.1
|
FBLN5
|
fibulin 5 |
| chr6_+_153552455 | 0.05 |
ENST00000392385.2
|
AL590867.1
|
Uncharacterized protein; cDNA FLJ59044, highly similar to LINE-1 reverse transcriptase homolog |
| chr3_-_114173530 | 0.05 |
ENST00000470311.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr15_+_76352178 | 0.05 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr17_+_61151306 | 0.05 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr14_-_51027838 | 0.05 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr21_-_35284635 | 0.05 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr6_-_109702885 | 0.05 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr7_-_84122033 | 0.05 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_+_210501589 | 0.04 |
ENST00000413764.2
ENST00000541565.1 |
HHAT
|
hedgehog acyltransferase |
| chr11_-_85430356 | 0.04 |
ENST00000526999.1
|
SYTL2
|
synaptotagmin-like 2 |
| chr5_+_176811431 | 0.04 |
ENST00000512593.1
ENST00000324417.5 |
SLC34A1
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr10_-_62060232 | 0.04 |
ENST00000503925.1
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr5_+_118668846 | 0.04 |
ENST00000513374.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr7_+_135611542 | 0.04 |
ENST00000416501.1
|
AC015987.2
|
AC015987.2 |
| chr5_+_133562095 | 0.04 |
ENST00000602919.1
|
CTD-2410N18.3
|
CTD-2410N18.3 |
| chrX_-_21676442 | 0.04 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr3_-_108248169 | 0.04 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr8_+_22424551 | 0.04 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr9_+_130026756 | 0.04 |
ENST00000314904.5
ENST00000373387.4 |
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr14_+_38065052 | 0.04 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chrX_+_100224676 | 0.04 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr8_+_107738240 | 0.04 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr11_-_121986923 | 0.04 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr7_-_77427676 | 0.04 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr3_-_72149553 | 0.04 |
ENST00000468646.2
ENST00000464271.1 |
LINC00877
|
long intergenic non-protein coding RNA 877 |
| chr18_-_53253000 | 0.04 |
ENST00000566514.1
|
TCF4
|
transcription factor 4 |
| chr4_+_71019903 | 0.04 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr3_+_130569429 | 0.04 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr16_+_72459838 | 0.04 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr18_+_57567180 | 0.04 |
ENST00000316660.6
ENST00000269518.9 |
PMAIP1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr10_-_50970382 | 0.04 |
ENST00000419399.1
ENST00000432695.1 |
OGDHL
|
oxoglutarate dehydrogenase-like |
| chrX_-_106243451 | 0.04 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr13_-_67802549 | 0.04 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr2_-_224467002 | 0.04 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr7_+_55433131 | 0.04 |
ENST00000254770.2
|
LANCL2
|
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr12_-_52946923 | 0.04 |
ENST00000267119.5
|
KRT71
|
keratin 71 |
| chr3_+_141106643 | 0.04 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr16_-_18887627 | 0.04 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr2_+_210444298 | 0.04 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr4_-_20985632 | 0.03 |
ENST00000359001.5
|
KCNIP4
|
Kv channel interacting protein 4 |
| chr6_-_97731019 | 0.03 |
ENST00000275053.4
|
MMS22L
|
MMS22-like, DNA repair protein |
| chr9_+_44867571 | 0.03 |
ENST00000377548.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr7_+_37723336 | 0.03 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr7_-_27169801 | 0.03 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr14_+_67831576 | 0.03 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr3_-_114477787 | 0.03 |
ENST00000464560.1
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr7_-_22233442 | 0.03 |
ENST00000401957.2
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr6_-_116833500 | 0.03 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr12_-_3862245 | 0.03 |
ENST00000252322.1
ENST00000440314.2 |
EFCAB4B
|
EF-hand calcium binding domain 4B |
| chr21_+_30673091 | 0.03 |
ENST00000447177.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr12_+_49621658 | 0.03 |
ENST00000541364.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr2_+_149447783 | 0.03 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr4_-_39979576 | 0.03 |
ENST00000303538.8
ENST00000503396.1 |
PDS5A
|
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of SHOX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.4 | GO:0001714 | endodermal cell fate specification(GO:0001714) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.2 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.1 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:1902748 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.0 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.0 | GO:0048193 | Golgi vesicle transport(GO:0048193) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |