Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
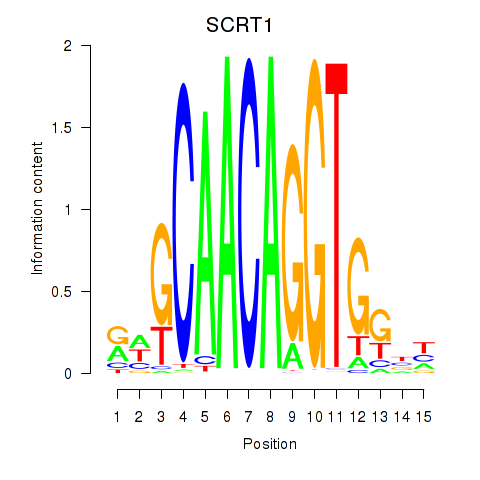
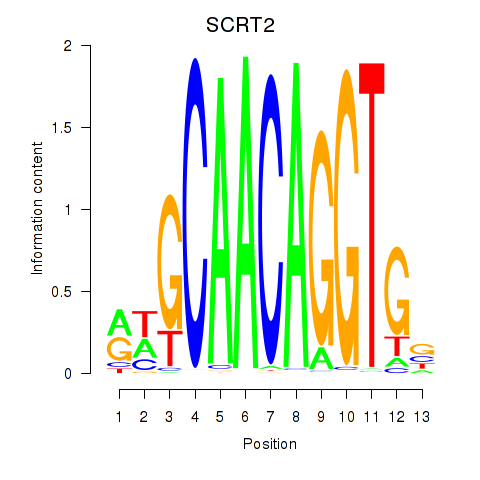
Results for SCRT1_SCRT2
Z-value: 0.60
Transcription factors associated with SCRT1_SCRT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SCRT1
|
ENSG00000170616.9 | scratch family transcriptional repressor 1 |
|
SCRT2
|
ENSG00000215397.3 | scratch family transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SCRT2 | hg19_v2_chr20_-_656823_656902 | -0.17 | 7.5e-01 | Click! |
| SCRT1 | hg19_v2_chr8_-_145559943_145559943 | -0.17 | 7.5e-01 | Click! |
Activity profile of SCRT1_SCRT2 motif
Sorted Z-values of SCRT1_SCRT2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_54317532 | 0.47 |
ENST00000418927.1
|
AL162759.1
|
AL162759.1 |
| chr17_-_15496722 | 0.46 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr6_-_83903600 | 0.31 |
ENST00000506587.1
ENST00000507554.1 |
PGM3
|
phosphoglucomutase 3 |
| chr1_+_33283246 | 0.29 |
ENST00000526230.1
ENST00000531256.1 ENST00000482212.1 |
S100PBP
|
S100P binding protein |
| chr1_+_109289279 | 0.29 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr8_-_33330595 | 0.28 |
ENST00000524021.1
ENST00000335589.3 |
FUT10
|
fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
| chr5_-_59064458 | 0.28 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr20_-_23402028 | 0.27 |
ENST00000398425.3
ENST00000432543.2 ENST00000377026.4 |
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta |
| chr3_-_143567262 | 0.26 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr2_+_149402009 | 0.26 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr7_+_12726623 | 0.25 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr2_-_24583314 | 0.25 |
ENST00000443927.1
ENST00000406921.3 ENST00000412011.1 |
ITSN2
|
intersectin 2 |
| chr8_-_133772870 | 0.25 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr2_-_24583583 | 0.24 |
ENST00000355123.4
|
ITSN2
|
intersectin 2 |
| chr6_-_27440837 | 0.23 |
ENST00000211936.6
|
ZNF184
|
zinc finger protein 184 |
| chr5_-_73936544 | 0.23 |
ENST00000509127.2
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr6_-_27440460 | 0.23 |
ENST00000377419.1
|
ZNF184
|
zinc finger protein 184 |
| chr12_-_31478428 | 0.23 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr15_-_76352069 | 0.22 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr1_-_222885770 | 0.22 |
ENST00000355727.2
ENST00000340020.6 |
AIDA
|
axin interactor, dorsalization associated |
| chr2_-_110962544 | 0.22 |
ENST00000355301.4
ENST00000445609.2 ENST00000417665.1 ENST00000418527.1 ENST00000316534.4 ENST00000393272.3 |
NPHP1
|
nephronophthisis 1 (juvenile) |
| chr2_+_18059906 | 0.21 |
ENST00000304101.4
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr8_-_110988070 | 0.20 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chr3_+_167453026 | 0.20 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr19_-_409134 | 0.19 |
ENST00000332235.6
|
C2CD4C
|
C2 calcium-dependent domain containing 4C |
| chr16_+_1031762 | 0.18 |
ENST00000293894.3
|
SOX8
|
SRY (sex determining region Y)-box 8 |
| chr9_+_69650263 | 0.17 |
ENST00000322495.3
|
AL445665.1
|
Protein LOC100996643 |
| chr18_+_52495426 | 0.17 |
ENST00000262094.5
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr1_+_33283043 | 0.17 |
ENST00000373476.1
ENST00000373475.5 ENST00000529027.1 ENST00000398243.3 |
S100PBP
|
S100P binding protein |
| chr4_-_6675550 | 0.17 |
ENST00000513179.1
ENST00000515205.1 |
RP11-539L10.3
|
RP11-539L10.3 |
| chr1_+_222791417 | 0.16 |
ENST00000344922.5
ENST00000344441.6 ENST00000344507.1 |
MIA3
|
melanoma inhibitory activity family, member 3 |
| chr3_+_141144963 | 0.15 |
ENST00000510726.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr18_+_47088401 | 0.15 |
ENST00000261292.4
ENST00000427224.2 ENST00000580036.1 |
LIPG
|
lipase, endothelial |
| chrX_-_55057403 | 0.15 |
ENST00000396198.3
ENST00000335854.4 ENST00000455688.1 ENST00000330807.5 |
ALAS2
|
aminolevulinate, delta-, synthase 2 |
| chr4_-_119757239 | 0.15 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr14_+_67708137 | 0.14 |
ENST00000556345.1
ENST00000555925.1 ENST00000557783.1 |
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr2_-_24583168 | 0.14 |
ENST00000361999.3
|
ITSN2
|
intersectin 2 |
| chr5_-_114631958 | 0.14 |
ENST00000395557.4
|
CCDC112
|
coiled-coil domain containing 112 |
| chr17_-_45056606 | 0.13 |
ENST00000322329.3
|
RPRML
|
reprimo-like |
| chr10_+_99894399 | 0.13 |
ENST00000298999.3
ENST00000314594.5 |
R3HCC1L
|
R3H domain and coiled-coil containing 1-like |
| chr4_-_141348789 | 0.13 |
ENST00000414773.1
|
CLGN
|
calmegin |
| chr9_+_74526532 | 0.13 |
ENST00000486911.2
|
C9orf85
|
chromosome 9 open reading frame 85 |
| chr12_-_67197760 | 0.13 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr17_+_63133587 | 0.12 |
ENST00000449996.3
ENST00000262406.9 |
RGS9
|
regulator of G-protein signaling 9 |
| chr6_+_71123107 | 0.12 |
ENST00000370479.3
ENST00000505769.1 ENST00000515323.1 ENST00000515280.1 ENST00000507085.1 ENST00000457062.2 ENST00000361499.3 |
FAM135A
|
family with sequence similarity 135, member A |
| chr4_+_120133791 | 0.12 |
ENST00000274030.6
|
USP53
|
ubiquitin specific peptidase 53 |
| chr17_+_14277419 | 0.11 |
ENST00000436469.1
|
AC022816.2
|
AC022816.2 |
| chrX_-_99986494 | 0.11 |
ENST00000372989.1
ENST00000455616.1 ENST00000454200.2 ENST00000276141.6 |
SYTL4
|
synaptotagmin-like 4 |
| chr16_-_50402690 | 0.11 |
ENST00000394689.2
|
BRD7
|
bromodomain containing 7 |
| chr5_-_114632307 | 0.11 |
ENST00000506442.1
ENST00000379611.5 |
CCDC112
|
coiled-coil domain containing 112 |
| chr1_-_219615984 | 0.11 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr16_-_29757272 | 0.11 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr11_-_85430163 | 0.11 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr5_+_138611798 | 0.10 |
ENST00000502394.1
|
MATR3
|
matrin 3 |
| chr9_+_100174344 | 0.10 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr19_-_49250054 | 0.10 |
ENST00000602105.1
ENST00000332955.2 |
IZUMO1
|
izumo sperm-egg fusion 1 |
| chr5_-_73936451 | 0.10 |
ENST00000537006.1
|
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chrY_-_21239221 | 0.10 |
ENST00000447937.1
ENST00000331787.2 |
TTTY14
|
testis-specific transcript, Y-linked 14 (non-protein coding) |
| chr2_+_10861775 | 0.10 |
ENST00000272238.4
ENST00000381661.3 |
ATP6V1C2
|
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2 |
| chr2_+_223289208 | 0.10 |
ENST00000321276.7
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr12_+_96337061 | 0.10 |
ENST00000266736.2
|
AMDHD1
|
amidohydrolase domain containing 1 |
| chr11_-_85430204 | 0.10 |
ENST00000389958.3
ENST00000527794.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr16_+_67063262 | 0.10 |
ENST00000565389.1
|
CBFB
|
core-binding factor, beta subunit |
| chr1_+_222988464 | 0.10 |
ENST00000420335.1
|
RP11-452F19.3
|
RP11-452F19.3 |
| chr1_+_55271736 | 0.10 |
ENST00000358193.3
ENST00000371273.3 |
C1orf177
|
chromosome 1 open reading frame 177 |
| chr9_+_74526384 | 0.10 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr3_+_167453493 | 0.10 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr3_-_49851313 | 0.10 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr12_-_21810765 | 0.10 |
ENST00000450584.1
ENST00000350669.1 |
LDHB
|
lactate dehydrogenase B |
| chr16_+_67063855 | 0.10 |
ENST00000563939.2
|
CBFB
|
core-binding factor, beta subunit |
| chr3_-_16554881 | 0.10 |
ENST00000451036.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr20_+_5987890 | 0.09 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr3_-_42845951 | 0.09 |
ENST00000418900.2
ENST00000430190.1 |
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr4_-_90757364 | 0.09 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr21_-_35987438 | 0.09 |
ENST00000313806.4
|
RCAN1
|
regulator of calcineurin 1 |
| chr17_+_48351785 | 0.09 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr15_+_63889577 | 0.09 |
ENST00000534939.1
ENST00000539570.3 |
FBXL22
|
F-box and leucine-rich repeat protein 22 |
| chr12_-_21810726 | 0.09 |
ENST00000396076.1
|
LDHB
|
lactate dehydrogenase B |
| chr3_-_42846021 | 0.09 |
ENST00000321331.7
|
HIGD1A
|
HIG1 hypoxia inducible domain family, member 1A |
| chr3_+_16216137 | 0.09 |
ENST00000339732.5
|
GALNT15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr11_+_44748361 | 0.09 |
ENST00000533202.1
ENST00000533080.1 ENST00000520358.2 ENST00000520999.2 |
TSPAN18
|
tetraspanin 18 |
| chrX_+_17653413 | 0.09 |
ENST00000398097.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr2_+_187350973 | 0.09 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr16_+_2286726 | 0.08 |
ENST00000382437.4
ENST00000569184.1 |
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chr8_-_91657740 | 0.08 |
ENST00000422900.1
|
TMEM64
|
transmembrane protein 64 |
| chr13_+_76362974 | 0.08 |
ENST00000497947.2
|
LMO7
|
LIM domain 7 |
| chr1_+_67395922 | 0.08 |
ENST00000401042.3
ENST00000355356.3 |
MIER1
|
mesoderm induction early response 1, transcriptional regulator |
| chr4_-_119757322 | 0.08 |
ENST00000379735.5
|
SEC24D
|
SEC24 family member D |
| chr3_+_184056614 | 0.08 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr12_-_33049690 | 0.08 |
ENST00000070846.6
ENST00000340811.4 |
PKP2
|
plakophilin 2 |
| chr16_-_53537105 | 0.08 |
ENST00000568596.1
ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP
|
AKT interacting protein |
| chr17_-_10101868 | 0.08 |
ENST00000432992.2
ENST00000540214.1 |
GAS7
|
growth arrest-specific 7 |
| chr7_+_94536898 | 0.08 |
ENST00000433360.1
ENST00000340694.4 ENST00000424654.1 |
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr2_+_187350883 | 0.08 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr12_+_50794730 | 0.08 |
ENST00000523389.1
ENST00000518561.1 ENST00000347328.5 ENST00000550260.1 |
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr22_+_22723969 | 0.08 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr12_+_16500037 | 0.08 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr15_+_84904525 | 0.08 |
ENST00000510439.2
|
GOLGA6L4
|
golgin A6 family-like 4 |
| chr17_+_53343577 | 0.08 |
ENST00000573945.1
|
HLF
|
hepatic leukemia factor |
| chr14_+_67707826 | 0.08 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr1_+_151483855 | 0.08 |
ENST00000427934.2
ENST00000271636.7 |
CGN
|
cingulin |
| chr17_+_63133526 | 0.08 |
ENST00000443584.3
|
RGS9
|
regulator of G-protein signaling 9 |
| chr3_-_96337000 | 0.08 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr10_+_96953957 | 0.08 |
ENST00000341686.3
ENST00000430183.1 |
C10orf129
|
chromosome 10 open reading frame 129 |
| chr7_+_12727250 | 0.08 |
ENST00000404894.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr4_-_170897045 | 0.07 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr8_-_82644562 | 0.07 |
ENST00000520604.1
ENST00000521742.1 ENST00000520635.1 |
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr4_-_70080449 | 0.07 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr5_+_74011328 | 0.07 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr1_-_95392635 | 0.07 |
ENST00000538964.1
ENST00000394202.4 ENST00000370206.4 |
CNN3
|
calponin 3, acidic |
| chr1_-_9129735 | 0.07 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr10_+_99894380 | 0.07 |
ENST00000370584.3
|
R3HCC1L
|
R3H domain and coiled-coil containing 1-like |
| chr5_+_148651409 | 0.07 |
ENST00000296721.4
|
AFAP1L1
|
actin filament associated protein 1-like 1 |
| chr3_+_141144954 | 0.07 |
ENST00000441582.2
ENST00000321464.5 |
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr18_+_47087390 | 0.07 |
ENST00000583083.1
|
LIPG
|
lipase, endothelial |
| chr17_+_19920456 | 0.07 |
ENST00000582604.1
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr12_-_64784306 | 0.07 |
ENST00000543259.1
|
C12orf56
|
chromosome 12 open reading frame 56 |
| chr18_-_10791648 | 0.07 |
ENST00000583325.1
|
PIEZO2
|
piezo-type mechanosensitive ion channel component 2 |
| chr11_-_8816375 | 0.07 |
ENST00000530580.1
|
ST5
|
suppression of tumorigenicity 5 |
| chrX_+_55744228 | 0.07 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr16_-_84076241 | 0.07 |
ENST00000568178.1
|
SLC38A8
|
solute carrier family 38, member 8 |
| chr14_+_68086515 | 0.07 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr8_-_133772794 | 0.06 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr8_-_91657909 | 0.06 |
ENST00000418210.2
|
TMEM64
|
transmembrane protein 64 |
| chr10_-_60027642 | 0.06 |
ENST00000373935.3
|
IPMK
|
inositol polyphosphate multikinase |
| chr6_+_7107830 | 0.06 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr1_-_104239076 | 0.06 |
ENST00000370080.3
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr3_+_87276407 | 0.06 |
ENST00000471660.1
ENST00000263780.4 ENST00000494980.1 |
CHMP2B
|
charged multivesicular body protein 2B |
| chr10_-_101945771 | 0.06 |
ENST00000370408.2
ENST00000407654.3 |
ERLIN1
|
ER lipid raft associated 1 |
| chr14_+_23776167 | 0.06 |
ENST00000554635.1
ENST00000557008.1 |
BCL2L2
BCL2L2-PABPN1
|
BCL2-like 2 BCL2L2-PABPN1 readthrough |
| chr1_+_1266654 | 0.06 |
ENST00000339381.5
|
TAS1R3
|
taste receptor, type 1, member 3 |
| chr13_+_20268547 | 0.06 |
ENST00000601204.1
|
AL354808.2
|
AL354808.2 |
| chr17_+_67410832 | 0.06 |
ENST00000590474.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_-_9129631 | 0.05 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr8_+_132952112 | 0.05 |
ENST00000520362.1
ENST00000519656.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr11_+_32112431 | 0.05 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr4_+_89378261 | 0.05 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr12_-_75905374 | 0.05 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr22_+_44577237 | 0.05 |
ENST00000415224.1
ENST00000417767.1 |
PARVG
|
parvin, gamma |
| chr6_+_71122974 | 0.05 |
ENST00000418814.2
|
FAM135A
|
family with sequence similarity 135, member A |
| chr1_+_215256467 | 0.05 |
ENST00000391894.2
ENST00000444842.2 |
KCNK2
|
potassium channel, subfamily K, member 2 |
| chr16_-_82045049 | 0.05 |
ENST00000532128.1
ENST00000328945.5 |
SDR42E1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr2_+_24397930 | 0.05 |
ENST00000295150.3
|
FAM228A
|
family with sequence similarity 228, member A |
| chr12_-_56221330 | 0.05 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chr14_-_102829051 | 0.05 |
ENST00000536961.2
ENST00000541568.2 ENST00000216756.6 |
CINP
|
cyclin-dependent kinase 2 interacting protein |
| chr1_-_104238912 | 0.05 |
ENST00000330330.5
|
AMY1B
|
amylase, alpha 1B (salivary) |
| chr11_+_125461607 | 0.05 |
ENST00000527606.1
|
STT3A
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr13_+_49551020 | 0.05 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr4_-_52904425 | 0.05 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr3_+_46742823 | 0.05 |
ENST00000326431.3
|
TMIE
|
transmembrane inner ear |
| chr1_-_52456352 | 0.05 |
ENST00000371655.3
|
RAB3B
|
RAB3B, member RAS oncogene family |
| chr2_+_173600671 | 0.05 |
ENST00000409036.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_203499901 | 0.05 |
ENST00000303116.6
ENST00000392238.2 |
FAM117B
|
family with sequence similarity 117, member B |
| chr5_+_173763250 | 0.05 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr17_+_7461613 | 0.04 |
ENST00000438470.1
ENST00000436057.1 |
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr4_-_140223670 | 0.04 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr3_-_196910721 | 0.04 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr1_+_225965518 | 0.04 |
ENST00000304786.7
ENST00000366839.4 ENST00000366838.1 |
SRP9
|
signal recognition particle 9kDa |
| chr3_-_97690931 | 0.04 |
ENST00000360258.4
|
MINA
|
MYC induced nuclear antigen |
| chr2_-_119916459 | 0.04 |
ENST00000272520.3
|
C1QL2
|
complement component 1, q subcomponent-like 2 |
| chr10_+_90672113 | 0.04 |
ENST00000371922.1
|
STAMBPL1
|
STAM binding protein-like 1 |
| chr5_+_148651469 | 0.04 |
ENST00000515000.1
|
AFAP1L1
|
actin filament associated protein 1-like 1 |
| chr16_+_53164956 | 0.04 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr6_+_7107999 | 0.04 |
ENST00000491191.1
ENST00000379938.2 ENST00000471433.1 |
RREB1
|
ras responsive element binding protein 1 |
| chr21_-_34852304 | 0.04 |
ENST00000542230.2
|
TMEM50B
|
transmembrane protein 50B |
| chr2_-_241759622 | 0.04 |
ENST00000320389.7
ENST00000498729.2 |
KIF1A
|
kinesin family member 1A |
| chr5_+_138089100 | 0.04 |
ENST00000520339.1
ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr3_+_133465228 | 0.04 |
ENST00000482271.1
ENST00000264998.3 |
TF
|
transferrin |
| chr6_+_7108210 | 0.04 |
ENST00000467782.1
ENST00000334984.6 ENST00000349384.6 |
RREB1
|
ras responsive element binding protein 1 |
| chr10_+_35484053 | 0.04 |
ENST00000487763.1
ENST00000473940.1 ENST00000488328.1 ENST00000356917.5 |
CREM
|
cAMP responsive element modulator |
| chr3_+_107096188 | 0.04 |
ENST00000261058.1
|
CCDC54
|
coiled-coil domain containing 54 |
| chr8_+_82644669 | 0.04 |
ENST00000297265.4
|
CHMP4C
|
charged multivesicular body protein 4C |
| chr12_+_50794891 | 0.04 |
ENST00000517559.1
|
LARP4
|
La ribonucleoprotein domain family, member 4 |
| chr2_+_173600514 | 0.04 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr2_+_61244697 | 0.04 |
ENST00000401576.1
ENST00000295030.5 ENST00000414712.2 |
PEX13
|
peroxisomal biogenesis factor 13 |
| chr13_-_25746416 | 0.04 |
ENST00000515384.1
ENST00000357816.2 |
AMER2
|
APC membrane recruitment protein 2 |
| chr11_+_34654011 | 0.04 |
ENST00000531794.1
|
EHF
|
ets homologous factor |
| chr2_+_173600565 | 0.04 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr9_+_215158 | 0.03 |
ENST00000479404.1
|
DOCK8
|
dedicator of cytokinesis 8 |
| chr1_-_247335269 | 0.03 |
ENST00000543802.2
ENST00000491356.1 ENST00000472531.1 ENST00000340684.6 |
ZNF124
|
zinc finger protein 124 |
| chrX_+_13707235 | 0.03 |
ENST00000464506.1
|
RAB9A
|
RAB9A, member RAS oncogene family |
| chr7_+_130126165 | 0.03 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr2_-_178753465 | 0.03 |
ENST00000389683.3
|
PDE11A
|
phosphodiesterase 11A |
| chr1_-_156307992 | 0.03 |
ENST00000415548.1
|
CCT3
|
chaperonin containing TCP1, subunit 3 (gamma) |
| chr3_+_16216210 | 0.03 |
ENST00000437509.1
|
GALNT15
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr22_-_30662828 | 0.03 |
ENST00000403463.1
ENST00000215781.2 |
OSM
|
oncostatin M |
| chr16_-_30022293 | 0.03 |
ENST00000565273.1
ENST00000567332.2 ENST00000350119.4 |
DOC2A
|
double C2-like domains, alpha |
| chr19_+_30097181 | 0.03 |
ENST00000586420.1
ENST00000221770.3 ENST00000392279.3 ENST00000590688.1 |
POP4
|
processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr6_-_170124027 | 0.03 |
ENST00000366780.4
ENST00000339209.4 |
PHF10
|
PHD finger protein 10 |
| chr5_-_127873496 | 0.03 |
ENST00000508989.1
|
FBN2
|
fibrillin 2 |
| chr1_-_225615599 | 0.03 |
ENST00000421383.1
ENST00000272163.4 |
LBR
|
lamin B receptor |
| chr17_-_73150599 | 0.03 |
ENST00000392566.2
ENST00000581874.1 |
HN1
|
hematological and neurological expressed 1 |
| chr19_-_51220176 | 0.03 |
ENST00000359082.3
ENST00000293441.1 |
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chrX_+_118370211 | 0.03 |
ENST00000217971.7
|
PGRMC1
|
progesterone receptor membrane component 1 |
| chr15_+_80351910 | 0.03 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr4_-_149363662 | 0.03 |
ENST00000355292.3
ENST00000358102.3 |
NR3C2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr8_-_82645082 | 0.03 |
ENST00000523361.1
|
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr1_-_21044489 | 0.03 |
ENST00000247986.2
|
KIF17
|
kinesin family member 17 |
| chr5_+_15500280 | 0.03 |
ENST00000504595.1
|
FBXL7
|
F-box and leucine-rich repeat protein 7 |
| chr19_+_12862604 | 0.02 |
ENST00000553030.1
|
BEST2
|
bestrophin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of SCRT1_SCRT2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.2 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.1 | 0.2 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.1 | 0.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.3 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.3 | GO:0022615 | protein to membrane docking(GO:0022615) |
| 0.0 | 0.3 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:1990539 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.2 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.3 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.0 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.0 | 0.0 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |