Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
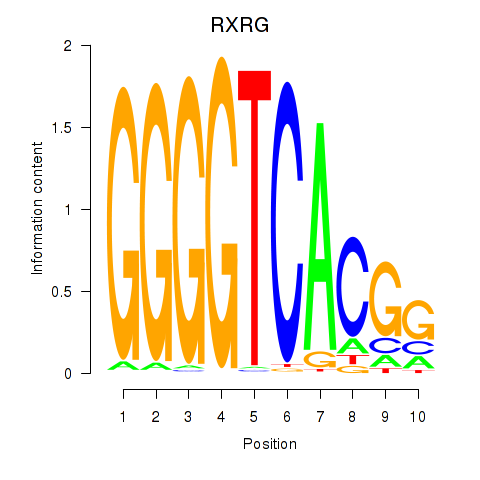
Results for RXRG
Z-value: 2.17
Transcription factors associated with RXRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRG
|
ENSG00000143171.8 | retinoid X receptor gamma |
Activity profile of RXRG motif
Sorted Z-values of RXRG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_74212073 | 2.35 |
ENST00000441217.1
|
AC073046.25
|
AC073046.25 |
| chr2_+_105471969 | 1.41 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr15_+_41221536 | 1.40 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chr6_-_34664612 | 1.19 |
ENST00000374023.3
ENST00000374026.3 |
C6orf106
|
chromosome 6 open reading frame 106 |
| chr1_+_202317855 | 1.17 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr1_-_183538319 | 1.10 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr19_-_1174226 | 1.06 |
ENST00000587024.1
ENST00000361757.3 |
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr4_-_90758227 | 1.03 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_+_7211280 | 1.02 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chrX_-_16888276 | 1.01 |
ENST00000493145.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr7_-_35734176 | 1.00 |
ENST00000413517.1
ENST00000438224.1 |
HERPUD2
|
HERPUD family member 2 |
| chr2_+_242913327 | 0.96 |
ENST00000426962.1
|
AC093642.3
|
AC093642.3 |
| chr17_+_3627185 | 0.96 |
ENST00000325418.4
|
GSG2
|
germ cell associated 2 (haspin) |
| chr16_+_71660052 | 0.94 |
ENST00000567566.1
|
MARVELD3
|
MARVEL domain containing 3 |
| chr6_+_35227449 | 0.93 |
ENST00000373953.3
ENST00000440666.2 ENST00000339411.5 |
ZNF76
|
zinc finger protein 76 |
| chr11_-_47447970 | 0.90 |
ENST00000298852.3
ENST00000530912.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr19_+_6361841 | 0.90 |
ENST00000596605.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr17_-_72889697 | 0.90 |
ENST00000310226.6
|
FADS6
|
fatty acid desaturase 6 |
| chr17_+_47074758 | 0.90 |
ENST00000290341.3
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr9_-_131534188 | 0.87 |
ENST00000414921.1
|
ZER1
|
zyg-11 related, cell cycle regulator |
| chr12_+_57998400 | 0.87 |
ENST00000548804.1
ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3
|
deltex homolog 3 (Drosophila) |
| chr19_+_52873166 | 0.87 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr17_+_59477233 | 0.87 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chr12_-_120806960 | 0.86 |
ENST00000257552.2
|
MSI1
|
musashi RNA-binding protein 1 |
| chr17_-_43568062 | 0.83 |
ENST00000421073.2
ENST00000584420.1 ENST00000589780.1 ENST00000430334.3 |
PLEKHM1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr6_-_32122106 | 0.83 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr11_+_65122216 | 0.82 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr3_+_14989076 | 0.82 |
ENST00000413118.1
ENST00000425241.1 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr16_+_70207686 | 0.81 |
ENST00000541793.2
ENST00000314151.8 ENST00000565806.1 ENST00000569347.2 ENST00000536907.2 |
CLEC18C
|
C-type lectin domain family 18, member C |
| chr17_+_47075023 | 0.81 |
ENST00000431824.2
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr11_-_47447767 | 0.80 |
ENST00000530651.1
ENST00000524447.2 ENST00000531051.2 ENST00000526993.1 ENST00000602866.1 |
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3 |
| chr1_+_202317815 | 0.79 |
ENST00000608999.1
ENST00000336894.4 ENST00000480184.1 |
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr14_+_38065052 | 0.79 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr11_-_65430251 | 0.79 |
ENST00000534283.1
ENST00000527749.1 ENST00000533187.1 ENST00000525693.1 ENST00000534558.1 ENST00000532879.1 ENST00000532999.1 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr19_+_50354430 | 0.78 |
ENST00000599732.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr17_-_73844722 | 0.78 |
ENST00000586257.1
|
WBP2
|
WW domain binding protein 2 |
| chr22_-_29784519 | 0.78 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr12_-_98897617 | 0.78 |
ENST00000501499.1
|
RP11-181C3.1
|
Uncharacterized protein |
| chr10_-_81203972 | 0.78 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr17_+_36584662 | 0.77 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr11_-_65429891 | 0.77 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr1_+_109642799 | 0.76 |
ENST00000602755.1
|
SCARNA2
|
small Cajal body-specific RNA 2 |
| chr19_-_14117074 | 0.76 |
ENST00000588885.1
ENST00000254325.4 |
RFX1
|
regulatory factor X, 1 (influences HLA class II expression) |
| chr17_+_7210898 | 0.75 |
ENST00000572815.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr22_-_30234218 | 0.75 |
ENST00000307790.3
ENST00000542393.1 ENST00000397771.2 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr6_+_24495185 | 0.74 |
ENST00000348925.2
|
ALDH5A1
|
aldehyde dehydrogenase 5 family, member A1 |
| chr22_-_43567750 | 0.73 |
ENST00000494035.1
|
TTLL12
|
tubulin tyrosine ligase-like family, member 12 |
| chr19_-_6424283 | 0.72 |
ENST00000595258.1
ENST00000595548.1 |
KHSRP
|
KH-type splicing regulatory protein |
| chr17_-_31404 | 0.72 |
ENST00000343572.7
|
DOC2B
|
double C2-like domains, beta |
| chr14_+_59100774 | 0.72 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chrX_-_30327495 | 0.72 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr2_-_121624973 | 0.71 |
ENST00000603720.1
|
RP11-297J22.1
|
RP11-297J22.1 |
| chr1_+_1243947 | 0.71 |
ENST00000379031.5
|
PUSL1
|
pseudouridylate synthase-like 1 |
| chr19_-_14228541 | 0.71 |
ENST00000590853.1
ENST00000308677.4 |
PRKACA
|
protein kinase, cAMP-dependent, catalytic, alpha |
| chr10_-_17496222 | 0.71 |
ENST00000377602.4
|
ST8SIA6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr19_+_50354462 | 0.70 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr7_-_767249 | 0.70 |
ENST00000403562.1
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr22_-_29663954 | 0.69 |
ENST00000216085.7
|
RHBDD3
|
rhomboid domain containing 3 |
| chr15_+_76352178 | 0.69 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr16_+_67700673 | 0.69 |
ENST00000403458.4
ENST00000602365.1 |
C16orf86
|
chromosome 16 open reading frame 86 |
| chr6_+_43737939 | 0.68 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chr3_+_52454971 | 0.68 |
ENST00000465863.1
|
PHF7
|
PHD finger protein 7 |
| chr19_+_9251052 | 0.68 |
ENST00000247956.6
ENST00000360385.3 |
ZNF317
|
zinc finger protein 317 |
| chr11_-_1587166 | 0.68 |
ENST00000331588.4
|
DUSP8
|
dual specificity phosphatase 8 |
| chr12_+_117176113 | 0.67 |
ENST00000319176.7
|
RNFT2
|
ring finger protein, transmembrane 2 |
| chr11_-_62439727 | 0.67 |
ENST00000528862.1
|
C11orf48
|
chromosome 11 open reading frame 48 |
| chr19_-_6279932 | 0.67 |
ENST00000252674.7
|
MLLT1
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr17_+_7211656 | 0.66 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr19_+_7587555 | 0.66 |
ENST00000601003.1
|
MCOLN1
|
mucolipin 1 |
| chr5_+_170846640 | 0.65 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr19_-_10047219 | 0.64 |
ENST00000264833.4
|
OLFM2
|
olfactomedin 2 |
| chrX_-_70474377 | 0.64 |
ENST00000373978.1
ENST00000373981.1 |
ZMYM3
|
zinc finger, MYM-type 3 |
| chr20_+_42295745 | 0.63 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chrX_-_49042778 | 0.63 |
ENST00000538114.1
ENST00000376310.3 ENST00000376317.3 ENST00000417014.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr3_-_50329990 | 0.62 |
ENST00000417626.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr19_+_50354393 | 0.61 |
ENST00000391842.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr11_+_65383227 | 0.61 |
ENST00000355703.3
|
PCNXL3
|
pecanex-like 3 (Drosophila) |
| chr2_+_27301435 | 0.60 |
ENST00000380320.4
|
EMILIN1
|
elastin microfibril interfacer 1 |
| chr3_-_50329835 | 0.60 |
ENST00000429673.2
|
IFRD2
|
interferon-related developmental regulator 2 |
| chr19_+_18303992 | 0.60 |
ENST00000599612.2
|
MPV17L2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr6_+_43738444 | 0.59 |
ENST00000324450.6
ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA
|
vascular endothelial growth factor A |
| chr1_-_153919128 | 0.59 |
ENST00000361217.4
|
DENND4B
|
DENN/MADD domain containing 4B |
| chrX_+_48687283 | 0.59 |
ENST00000338270.1
|
ERAS
|
ES cell expressed Ras |
| chr17_+_33307503 | 0.59 |
ENST00000378526.4
ENST00000585941.1 ENST00000262327.5 ENST00000592690.1 ENST00000585740.1 |
LIG3
|
ligase III, DNA, ATP-dependent |
| chr19_+_12780512 | 0.58 |
ENST00000242796.4
|
WDR83
|
WD repeat domain 83 |
| chr11_+_327171 | 0.58 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr11_+_46722368 | 0.58 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr12_-_56223363 | 0.58 |
ENST00000546957.1
|
DNAJC14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr19_-_50432711 | 0.58 |
ENST00000597723.1
ENST00000599788.1 ENST00000596217.1 ENST00000593652.1 ENST00000599567.1 ENST00000600935.1 ENST00000596011.1 ENST00000596022.1 ENST00000597295.1 |
NUP62
IL4I1
|
nucleoporin 62kDa interleukin 4 induced 1 |
| chrX_-_47509994 | 0.58 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chrX_+_153672468 | 0.58 |
ENST00000393600.3
|
FAM50A
|
family with sequence similarity 50, member A |
| chr9_-_110251836 | 0.58 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr17_-_7832753 | 0.56 |
ENST00000303790.2
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr16_+_20912114 | 0.56 |
ENST00000567954.1
|
LYRM1
|
LYR motif containing 1 |
| chr16_-_23724518 | 0.56 |
ENST00000457008.2
|
ERN2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr19_-_4124079 | 0.56 |
ENST00000394867.4
ENST00000262948.5 |
MAP2K2
|
mitogen-activated protein kinase kinase 2 |
| chr3_-_125655882 | 0.55 |
ENST00000340333.3
|
ALG1L
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase-like |
| chr9_-_140009130 | 0.55 |
ENST00000497375.1
ENST00000371579.2 |
DPP7
|
dipeptidyl-peptidase 7 |
| chr11_-_65430554 | 0.55 |
ENST00000308639.9
ENST00000406246.3 |
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr4_+_184826418 | 0.55 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr2_-_228028829 | 0.55 |
ENST00000396625.3
ENST00000329662.7 |
COL4A4
|
collagen, type IV, alpha 4 |
| chr10_+_88718397 | 0.55 |
ENST00000372017.3
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr19_-_50432654 | 0.54 |
ENST00000596680.1
ENST00000594673.1 ENST00000597029.1 |
NUP62
|
nucleoporin 62kDa |
| chr1_-_154946792 | 0.54 |
ENST00000412170.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr22_-_29663690 | 0.54 |
ENST00000406335.1
|
RHBDD3
|
rhomboid domain containing 3 |
| chr22_+_30821732 | 0.54 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr17_-_79829190 | 0.54 |
ENST00000581876.1
ENST00000584461.1 ENST00000583868.1 ENST00000400721.4 ENST00000269321.7 |
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr7_+_143078652 | 0.54 |
ENST00000354434.4
ENST00000449423.2 |
ZYX
|
zyxin |
| chr22_+_19705928 | 0.54 |
ENST00000383045.3
ENST00000438754.2 |
SEPT5
|
septin 5 |
| chr22_+_19701985 | 0.53 |
ENST00000455784.2
ENST00000406395.1 |
SEPT5
|
septin 5 |
| chr22_-_20104700 | 0.53 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr17_-_27278304 | 0.53 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr17_-_38256973 | 0.53 |
ENST00000246672.3
|
NR1D1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr19_-_39390350 | 0.53 |
ENST00000447739.1
ENST00000358931.5 ENST00000407552.1 |
SIRT2
|
sirtuin 2 |
| chr8_-_145331153 | 0.53 |
ENST00000377412.4
|
KM-PA-2
|
KM-PA-2 protein; Uncharacterized protein |
| chr6_+_42018614 | 0.52 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr19_+_18451439 | 0.52 |
ENST00000597431.2
|
PGPEP1
|
pyroglutamyl-peptidase I |
| chr3_-_46735155 | 0.52 |
ENST00000318962.4
|
ALS2CL
|
ALS2 C-terminal like |
| chr19_-_44097188 | 0.52 |
ENST00000594374.1
|
L34079.2
|
L34079.2 |
| chr19_-_39390440 | 0.52 |
ENST00000249396.7
ENST00000414941.1 ENST00000392081.2 |
SIRT2
|
sirtuin 2 |
| chr19_+_49535169 | 0.52 |
ENST00000474913.1
ENST00000359342.6 |
CGB2
|
chorionic gonadotropin, beta polypeptide 2 |
| chr17_-_27277615 | 0.51 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr19_+_50353944 | 0.51 |
ENST00000594151.1
ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1
|
prostate tumor overexpressed 1 |
| chr18_+_56530692 | 0.51 |
ENST00000588601.1
|
ZNF532
|
zinc finger protein 532 |
| chr11_-_46722117 | 0.51 |
ENST00000311956.4
|
ARHGAP1
|
Rho GTPase activating protein 1 |
| chr19_+_50887585 | 0.51 |
ENST00000440232.2
ENST00000601098.1 ENST00000599857.1 ENST00000593887.1 |
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr11_-_62380199 | 0.51 |
ENST00000419857.1
ENST00000394773.2 |
EML3
|
echinoderm microtubule associated protein like 3 |
| chr1_-_6761855 | 0.50 |
ENST00000426784.1
ENST00000377573.5 ENST00000377577.5 ENST00000294401.7 |
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11 |
| chr1_+_7844312 | 0.50 |
ENST00000377541.1
|
PER3
|
period circadian clock 3 |
| chr2_-_45165994 | 0.50 |
ENST00000444871.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr1_-_41487383 | 0.50 |
ENST00000302946.8
ENST00000372613.2 ENST00000439569.2 ENST00000397197.2 |
SLFNL1
|
schlafen-like 1 |
| chr15_-_40574787 | 0.50 |
ENST00000434396.1
|
ANKRD63
|
ankyrin repeat domain 63 |
| chr11_+_66036004 | 0.50 |
ENST00000311481.6
ENST00000527397.1 |
RAB1B
|
RAB1B, member RAS oncogene family |
| chr12_-_48214345 | 0.50 |
ENST00000433685.1
ENST00000447463.1 ENST00000427332.2 |
HDAC7
|
histone deacetylase 7 |
| chr17_+_36905613 | 0.50 |
ENST00000539023.1
|
CTB-58E17.5
|
Uncharacterized protein; cDNA FLJ52623 |
| chr4_+_159690218 | 0.49 |
ENST00000264433.6
|
FNIP2
|
folliculin interacting protein 2 |
| chr2_+_27593389 | 0.49 |
ENST00000233575.2
ENST00000543024.1 ENST00000537606.1 |
SNX17
|
sorting nexin 17 |
| chr1_-_161015663 | 0.49 |
ENST00000534633.1
|
USF1
|
upstream transcription factor 1 |
| chr17_+_7210921 | 0.49 |
ENST00000573542.1
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chrX_+_48911090 | 0.49 |
ENST00000597275.1
|
CCDC120
|
coiled-coil domain containing 120 |
| chr1_-_212873267 | 0.49 |
ENST00000243440.1
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr12_-_6579833 | 0.49 |
ENST00000396308.3
|
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr22_-_32058166 | 0.49 |
ENST00000435900.1
ENST00000336566.4 |
PISD
|
phosphatidylserine decarboxylase |
| chr19_+_7968728 | 0.49 |
ENST00000397981.3
ENST00000545011.1 ENST00000397983.3 ENST00000397979.3 |
MAP2K7
|
mitogen-activated protein kinase kinase 7 |
| chr19_+_6361795 | 0.49 |
ENST00000596149.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr3_-_121467983 | 0.48 |
ENST00000472475.1
|
GOLGB1
|
golgin B1 |
| chr2_-_220117867 | 0.48 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr3_-_197463590 | 0.48 |
ENST00000296343.5
ENST00000389665.5 ENST00000449205.1 |
KIAA0226
|
KIAA0226 |
| chr17_+_7748233 | 0.48 |
ENST00000570632.1
|
KDM6B
|
lysine (K)-specific demethylase 6B |
| chr7_-_99717463 | 0.48 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr16_-_70719925 | 0.48 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr19_+_6361754 | 0.48 |
ENST00000597326.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr19_+_36203830 | 0.48 |
ENST00000262630.3
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr6_+_42018251 | 0.48 |
ENST00000372978.3
ENST00000494547.1 ENST00000456846.2 ENST00000372982.4 ENST00000472818.1 ENST00000372977.3 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr19_+_54705025 | 0.48 |
ENST00000441429.1
|
RPS9
|
ribosomal protein S9 |
| chr19_+_35759824 | 0.48 |
ENST00000343550.5
|
USF2
|
upstream transcription factor 2, c-fos interacting |
| chr12_-_71003568 | 0.48 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr2_+_237476419 | 0.48 |
ENST00000447924.1
|
ACKR3
|
atypical chemokine receptor 3 |
| chr20_-_2821271 | 0.47 |
ENST00000448755.1
ENST00000360652.2 |
PCED1A
|
PC-esterase domain containing 1A |
| chr20_-_4055812 | 0.47 |
ENST00000379526.1
|
RP11-352D3.2
|
Uncharacterized protein |
| chr5_-_132112921 | 0.47 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr2_-_242255117 | 0.47 |
ENST00000420451.1
ENST00000417540.1 ENST00000310931.4 |
HDLBP
|
high density lipoprotein binding protein |
| chr16_+_29985503 | 0.47 |
ENST00000543033.1
ENST00000279394.3 |
TAOK2
|
TAO kinase 2 |
| chr22_+_38093005 | 0.47 |
ENST00000406386.3
|
TRIOBP
|
TRIO and F-actin binding protein |
| chr7_-_766879 | 0.47 |
ENST00000537384.1
ENST00000417852.1 |
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr17_+_42264322 | 0.46 |
ENST00000446571.3
ENST00000357984.3 ENST00000538716.2 |
TMUB2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr1_-_154946825 | 0.46 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr17_+_79373540 | 0.46 |
ENST00000307745.7
|
RP11-1055B8.7
|
BAH and coiled-coil domain-containing protein 1 |
| chr11_-_45939565 | 0.46 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr7_+_143079000 | 0.46 |
ENST00000392910.2
|
ZYX
|
zyxin |
| chr19_-_1021113 | 0.46 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr3_-_119379427 | 0.45 |
ENST00000264231.3
ENST00000468801.1 ENST00000538678.1 |
POPDC2
|
popeye domain containing 2 |
| chr1_-_156460391 | 0.45 |
ENST00000360595.3
|
MEF2D
|
myocyte enhancer factor 2D |
| chr19_-_50432782 | 0.45 |
ENST00000413454.1
ENST00000596437.1 ENST00000341114.3 ENST00000595948.1 |
NUP62
IL4I1
|
nucleoporin 62kDa interleukin 4 induced 1 |
| chr1_-_51810778 | 0.45 |
ENST00000413473.2
ENST00000401051.3 ENST00000527205.1 |
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr7_+_99717230 | 0.44 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr10_-_135382876 | 0.44 |
ENST00000432597.2
ENST00000541506.1 |
SYCE1
SPRN
|
synaptonemal complex central element protein 1 shadow of prion protein homolog (zebrafish) |
| chr16_+_1401924 | 0.44 |
ENST00000204679.4
ENST00000529110.1 |
GNPTG
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit |
| chr7_-_35734607 | 0.44 |
ENST00000427455.1
|
HERPUD2
|
HERPUD family member 2 |
| chr11_-_46940074 | 0.44 |
ENST00000378623.1
ENST00000534404.1 |
LRP4
|
low density lipoprotein receptor-related protein 4 |
| chr9_-_123342415 | 0.44 |
ENST00000349780.4
ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr19_+_50432400 | 0.44 |
ENST00000423777.2
ENST00000600336.1 ENST00000597227.1 |
ATF5
|
activating transcription factor 5 |
| chr19_+_4402659 | 0.44 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chrX_-_47509887 | 0.43 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chr17_+_37793318 | 0.43 |
ENST00000336308.5
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr1_+_155146318 | 0.43 |
ENST00000368385.4
ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46
|
tripartite motif containing 46 |
| chr3_-_52486841 | 0.43 |
ENST00000496590.1
|
TNNC1
|
troponin C type 1 (slow) |
| chr1_+_230202936 | 0.43 |
ENST00000366672.4
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr11_+_72281681 | 0.43 |
ENST00000450804.3
|
RP11-169D4.1
|
RP11-169D4.1 |
| chr17_-_1531635 | 0.43 |
ENST00000571650.1
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr1_+_78245466 | 0.43 |
ENST00000477627.2
|
FAM73A
|
family with sequence similarity 73, member A |
| chr17_-_73511504 | 0.42 |
ENST00000581870.1
|
CASKIN2
|
CASK interacting protein 2 |
| chr1_+_160321120 | 0.42 |
ENST00000424754.1
|
NCSTN
|
nicastrin |
| chr1_+_23037323 | 0.42 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr12_-_56652111 | 0.42 |
ENST00000267116.7
|
ANKRD52
|
ankyrin repeat domain 52 |
| chr17_+_2240775 | 0.42 |
ENST00000268989.3
ENST00000426855.2 |
SGSM2
|
small G protein signaling modulator 2 |
| chr12_-_57634475 | 0.42 |
ENST00000393825.1
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
| chr21_-_46707793 | 0.42 |
ENST00000331343.7
ENST00000349485.5 |
POFUT2
|
protein O-fucosyltransferase 2 |
| chr5_+_133861790 | 0.42 |
ENST00000395003.1
|
PHF15
|
jade family PHD finger 2 |
| chr19_+_4304685 | 0.42 |
ENST00000601006.1
|
FSD1
|
fibronectin type III and SPRY domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RXRG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.5 | 1.5 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.4 | 1.3 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.4 | 3.3 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.4 | 1.4 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.3 | 1.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.3 | 0.9 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.3 | 1.6 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.3 | 1.6 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.3 | 0.8 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.2 | 1.7 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.2 | 0.2 | GO:1900200 | mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) |
| 0.2 | 1.4 | GO:0072069 | ascending thin limb development(GO:0072021) DCT cell differentiation(GO:0072069) metanephric ascending thin limb development(GO:0072218) metanephric DCT cell differentiation(GO:0072240) |
| 0.2 | 1.4 | GO:1903588 | regulation of neural retina development(GO:0061074) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.2 | 1.1 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.4 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.2 | 1.0 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) |
| 0.2 | 0.8 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.2 | 0.7 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 0.5 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.2 | 0.5 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.2 | 0.5 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.2 | 1.6 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.2 | 0.8 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.2 | 0.9 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.2 | 1.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.4 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.7 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.6 | GO:2000342 | regulation of muscle hyperplasia(GO:0014738) negative regulation of muscle hyperplasia(GO:0014740) negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.1 | 0.1 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.1 | 0.6 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.4 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 0.4 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.1 | 0.7 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.1 | 0.4 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.1 | 0.6 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.1 | 0.3 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.1 | 0.4 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.1 | 0.6 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.3 | GO:0060927 | Purkinje myocyte differentiation(GO:0003168) cardiac pacemaker cell fate commitment(GO:0060927) atrioventricular node cell fate commitment(GO:0060929) |
| 0.1 | 0.8 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.1 | 0.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.3 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.7 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.4 | GO:0042351 | 'de novo' GDP-L-fucose biosynthetic process(GO:0042351) |
| 0.1 | 0.3 | GO:0071879 | UDP-glucose catabolic process(GO:0006258) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.6 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.4 | GO:0051413 | response to cortisone(GO:0051413) |
| 0.1 | 0.7 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.5 | GO:1903911 | positive regulation of postsynaptic membrane organization(GO:1901628) positive regulation of receptor clustering(GO:1903911) |
| 0.1 | 0.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 1.0 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.1 | 0.4 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.4 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 0.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.1 | 0.7 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.3 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 1.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.1 | 0.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 1.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.9 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.1 | 0.5 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.1 | 0.3 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.8 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.3 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.5 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.5 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.2 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.1 | 0.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.1 | 1.0 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.2 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.4 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.5 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.1 | 0.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.3 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 0.2 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.1 | 0.7 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.3 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.4 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.2 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.1 | 1.4 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.3 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.1 | 0.6 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.1 | 1.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 1.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.7 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.6 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 | 0.3 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.2 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.1 | 0.2 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.1 | 0.3 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:1902822 | cleavage furrow ingression(GO:0036090) regulation of late endosome to lysosome transport(GO:1902822) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.4 | GO:0010918 | positive regulation of mitochondrial membrane potential(GO:0010918) |
| 0.0 | 0.2 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.4 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.4 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.7 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.3 | GO:1901216 | positive regulation of neuron death(GO:1901216) |
| 0.0 | 1.0 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.5 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.5 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.4 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.7 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.4 | GO:0009227 | UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 | 0.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.2 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.7 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.7 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.4 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:1903450 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.0 | 0.5 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.1 | GO:0000494 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.4 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.3 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.3 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.6 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.3 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.4 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) |
| 0.0 | 0.2 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.0 | 1.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0070318 | response to anoxia(GO:0034059) positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.5 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.4 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.2 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 1.6 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.2 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.4 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.3 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.2 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.5 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.3 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.2 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.1 | GO:0032796 | uropod organization(GO:0032796) |
| 0.0 | 0.8 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.7 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 1.0 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.4 | GO:0016573 | histone acetylation(GO:0016573) internal peptidyl-lysine acetylation(GO:0018393) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.5 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.3 | GO:1900825 | regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) |
| 0.0 | 0.4 | GO:0051438 | regulation of ubiquitin-protein transferase activity(GO:0051438) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0098712 | L-glutamate import across plasma membrane(GO:0098712) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0050434 | positive regulation of viral transcription(GO:0050434) |
| 0.0 | 0.2 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 1.0 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.5 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.4 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.4 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.3 | GO:1903020 | positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.7 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.4 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0090261 | positive regulation of inclusion body assembly(GO:0090261) |
| 0.0 | 0.0 | GO:0046984 | regulation of hemoglobin biosynthetic process(GO:0046984) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.6 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.5 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.0 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0032484 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.5 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.2 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.4 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.1 | GO:0036100 | leukotriene catabolic process(GO:0036100) leukotriene B4 catabolic process(GO:0036101) leukotriene B4 metabolic process(GO:0036102) icosanoid catabolic process(GO:1901523) fatty acid derivative catabolic process(GO:1901569) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.4 | 2.2 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.3 | 4.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 0.3 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.2 | 0.7 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.2 | 0.6 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.8 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 1.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.0 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.1 | 1.5 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 1.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.1 | 0.2 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.1 | 0.9 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.3 | GO:0043257 | laminin-8 complex(GO:0043257) |
| 0.1 | 1.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:1990742 | microvesicle(GO:1990742) |
| 0.1 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.5 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.1 | 0.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.5 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.5 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.4 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 1.1 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.5 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 1.0 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0097635 | Atg12-Atg5-Atg16 complex(GO:0034274) extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 2.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) symmetric synapse(GO:0032280) |
| 0.0 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.9 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.5 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.5 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 1.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.3 | 1.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.2 | 2.6 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 0.9 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.2 | 1.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.6 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.2 | 1.6 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.2 | 0.6 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.2 | 1.0 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 0.5 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.2 | 3.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 1.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.4 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.4 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.1 | 1.0 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.4 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.1 | 1.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.4 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.1 | 0.7 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.3 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.6 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.7 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.5 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 1.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.2 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.4 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.1 | 0.4 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.2 | GO:0052830 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.1 | 0.3 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.1 | 0.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 1.1 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 2.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 1.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 1.4 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) C-X-C chemokine binding(GO:0019958) |
| 0.0 | 1.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.6 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 1.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.5 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 1.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.0 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.6 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.6 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.7 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0097258 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 1.0 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0050659 | chondroitin 4-sulfotransferase activity(GO:0047756) N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.0 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 1.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 2.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.6 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.2 | 1.4 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.1 | 2.5 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.1 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 3.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 1.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.9 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 1.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.9 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 2.3 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.7 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.7 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |