Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
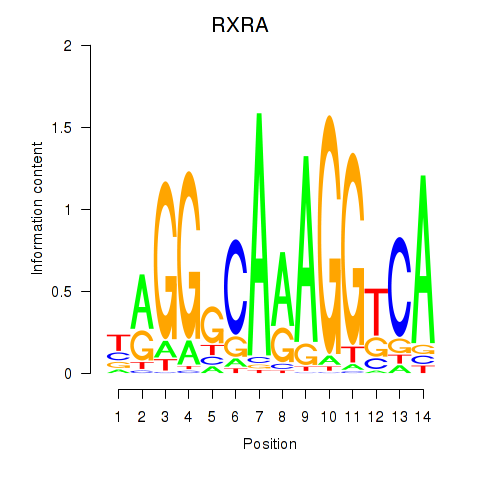
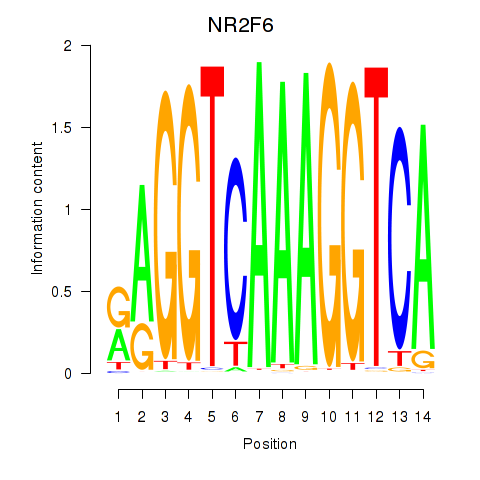
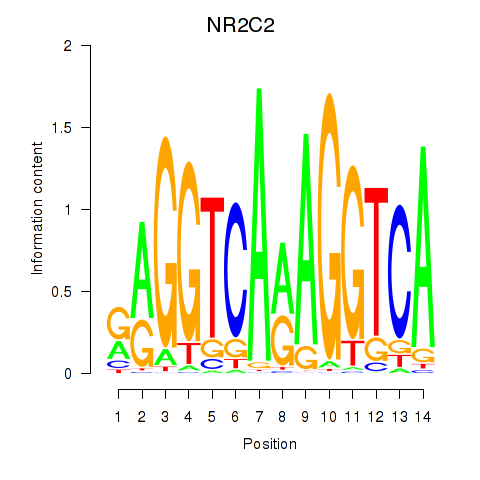
Results for RXRA_NR2F6_NR2C2
Z-value: 0.50
Transcription factors associated with RXRA_NR2F6_NR2C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRA
|
ENSG00000186350.8 | retinoid X receptor alpha |
|
NR2F6
|
ENSG00000160113.5 | nuclear receptor subfamily 2 group F member 6 |
|
NR2C2
|
ENSG00000177463.11 | nuclear receptor subfamily 2 group C member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F6 | hg19_v2_chr19_-_17356697_17356762 | 0.90 | 1.5e-02 | Click! |
| NR2C2 | hg19_v2_chr3_+_15045419_15045433 | 0.84 | 3.8e-02 | Click! |
| RXRA | hg19_v2_chr9_+_137298396_137298431 | 0.76 | 7.8e-02 | Click! |
Activity profile of RXRA_NR2F6_NR2C2 motif
Sorted Z-values of RXRA_NR2F6_NR2C2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_48867291 | 0.55 |
ENST00000435956.3
|
TMEM143
|
transmembrane protein 143 |
| chr6_+_143999185 | 0.39 |
ENST00000542769.1
ENST00000397980.3 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr19_+_39616410 | 0.36 |
ENST00000602004.1
ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr1_-_156675564 | 0.31 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr8_-_8639956 | 0.25 |
ENST00000522213.1
|
RP11-211C9.1
|
RP11-211C9.1 |
| chr16_+_30710462 | 0.24 |
ENST00000262518.4
ENST00000395059.2 ENST00000344771.4 |
SRCAP
|
Snf2-related CREBBP activator protein |
| chr16_+_81272287 | 0.21 |
ENST00000425577.2
ENST00000564552.1 |
BCMO1
|
beta-carotene 15,15'-monooxygenase 1 |
| chr7_+_143080063 | 0.21 |
ENST00000446634.1
|
ZYX
|
zyxin |
| chr19_-_36304201 | 0.20 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr2_+_73441350 | 0.20 |
ENST00000389501.4
|
SMYD5
|
SMYD family member 5 |
| chr7_+_97840739 | 0.19 |
ENST00000609256.1
|
BHLHA15
|
basic helix-loop-helix family, member a15 |
| chr8_-_135652051 | 0.19 |
ENST00000522257.1
|
ZFAT
|
zinc finger and AT hook domain containing |
| chr6_+_31926857 | 0.18 |
ENST00000375394.2
ENST00000544581.1 |
SKIV2L
|
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr9_+_104296163 | 0.18 |
ENST00000374819.2
ENST00000479306.1 |
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr17_+_4692230 | 0.17 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr7_-_75452673 | 0.17 |
ENST00000416943.1
|
CCL24
|
chemokine (C-C motif) ligand 24 |
| chr19_-_48867171 | 0.16 |
ENST00000377431.2
ENST00000436660.2 ENST00000541566.1 |
TMEM143
|
transmembrane protein 143 |
| chr2_+_159651821 | 0.16 |
ENST00000309950.3
ENST00000409042.1 |
DAPL1
|
death associated protein-like 1 |
| chr22_-_29784519 | 0.16 |
ENST00000357586.2
ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr11_-_111649015 | 0.16 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr3_+_9691117 | 0.16 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr11_-_111649074 | 0.15 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr20_+_44509857 | 0.15 |
ENST00000372523.1
ENST00000372520.1 |
ZSWIM1
|
zinc finger, SWIM-type containing 1 |
| chr3_+_190281229 | 0.15 |
ENST00000453359.1
|
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr7_-_74489609 | 0.15 |
ENST00000329959.4
ENST00000503250.2 ENST00000543840.1 |
WBSCR16
|
Williams-Beuren syndrome chromosome region 16 |
| chr2_+_10442993 | 0.15 |
ENST00000423674.1
ENST00000307845.3 |
HPCAL1
|
hippocalcin-like 1 |
| chr19_-_1021113 | 0.15 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr9_+_104296243 | 0.15 |
ENST00000466817.1
|
RNF20
|
ring finger protein 20, E3 ubiquitin protein ligase |
| chr22_+_47158518 | 0.14 |
ENST00000337137.4
ENST00000380995.1 ENST00000407381.3 |
TBC1D22A
|
TBC1 domain family, member 22A |
| chr12_+_57916584 | 0.14 |
ENST00000546632.1
ENST00000549623.1 ENST00000431731.2 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr5_-_176981417 | 0.13 |
ENST00000514747.1
ENST00000443375.2 ENST00000329540.5 |
FAM193B
|
family with sequence similarity 193, member B |
| chr16_+_30076052 | 0.13 |
ENST00000563987.1
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr16_+_2255841 | 0.12 |
ENST00000301725.7
|
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr12_-_109219937 | 0.12 |
ENST00000546697.1
|
SSH1
|
slingshot protein phosphatase 1 |
| chr9_-_16705069 | 0.12 |
ENST00000471301.2
|
BNC2
|
basonuclin 2 |
| chr12_+_132413739 | 0.12 |
ENST00000443358.2
|
PUS1
|
pseudouridylate synthase 1 |
| chr17_-_1613663 | 0.12 |
ENST00000330676.6
|
TLCD2
|
TLC domain containing 2 |
| chr21_-_43735628 | 0.12 |
ENST00000291525.10
ENST00000518498.1 |
TFF3
|
trefoil factor 3 (intestinal) |
| chr16_-_88752889 | 0.12 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr3_-_52002403 | 0.12 |
ENST00000490063.1
ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4
|
poly(rC) binding protein 4 |
| chr6_-_41747595 | 0.12 |
ENST00000373018.3
|
FRS3
|
fibroblast growth factor receptor substrate 3 |
| chr20_-_36661826 | 0.12 |
ENST00000373448.2
ENST00000373447.3 |
TTI1
|
TELO2 interacting protein 1 |
| chr11_-_45939374 | 0.11 |
ENST00000533151.1
ENST00000241041.3 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr11_+_118868830 | 0.11 |
ENST00000334418.1
|
CCDC84
|
coiled-coil domain containing 84 |
| chr16_-_67190152 | 0.11 |
ENST00000486556.1
|
TRADD
|
TNFRSF1A-associated via death domain |
| chr6_-_33771757 | 0.11 |
ENST00000507738.1
ENST00000266003.5 ENST00000430124.2 |
MLN
|
motilin |
| chr8_+_144816303 | 0.11 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr17_-_27503770 | 0.11 |
ENST00000533112.1
|
MYO18A
|
myosin XVIIIA |
| chr11_-_45939565 | 0.11 |
ENST00000525192.1
ENST00000378750.5 |
PEX16
|
peroxisomal biogenesis factor 16 |
| chr1_-_153513170 | 0.11 |
ENST00000368717.2
|
S100A5
|
S100 calcium binding protein A5 |
| chr5_-_33297946 | 0.11 |
ENST00000510327.1
|
CTD-2066L21.3
|
CTD-2066L21.3 |
| chrX_-_55020511 | 0.11 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr19_-_49339080 | 0.11 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr20_-_52687059 | 0.11 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr7_-_65447192 | 0.11 |
ENST00000421103.1
ENST00000345660.6 ENST00000304895.4 |
GUSB
|
glucuronidase, beta |
| chr1_-_154946792 | 0.11 |
ENST00000412170.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr17_-_71223839 | 0.11 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr22_+_47158578 | 0.11 |
ENST00000355704.3
|
TBC1D22A
|
TBC1 domain family, member 22A |
| chr1_-_155939437 | 0.11 |
ENST00000609707.1
|
ARHGEF2
|
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr22_-_20104700 | 0.11 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr1_-_17676070 | 0.11 |
ENST00000602074.1
|
AC004824.2
|
Uncharacterized protein |
| chr14_+_105331596 | 0.10 |
ENST00000556508.1
ENST00000414716.3 ENST00000453495.1 ENST00000418279.1 |
CEP170B
|
centrosomal protein 170B |
| chr19_+_44100632 | 0.10 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chrX_+_47083037 | 0.10 |
ENST00000523034.1
|
CDK16
|
cyclin-dependent kinase 16 |
| chr4_+_79475019 | 0.10 |
ENST00000508214.1
|
ANXA3
|
annexin A3 |
| chr17_-_42992856 | 0.10 |
ENST00000588316.1
ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP
|
glial fibrillary acidic protein |
| chr11_-_73687997 | 0.10 |
ENST00000545212.1
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr6_+_43737939 | 0.10 |
ENST00000372067.3
|
VEGFA
|
vascular endothelial growth factor A |
| chr19_-_17356697 | 0.10 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr9_-_140513231 | 0.10 |
ENST00000371417.3
|
C9orf37
|
chromosome 9 open reading frame 37 |
| chr7_-_83278322 | 0.10 |
ENST00000307792.3
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr2_+_219433281 | 0.10 |
ENST00000273064.6
ENST00000509807.2 ENST00000542068.1 |
RQCD1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr19_+_50919056 | 0.10 |
ENST00000599632.1
|
CTD-2545M3.6
|
CTD-2545M3.6 |
| chr16_-_4401284 | 0.10 |
ENST00000318059.3
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr1_+_197881592 | 0.09 |
ENST00000367391.1
ENST00000367390.3 |
LHX9
|
LIM homeobox 9 |
| chr19_+_44100727 | 0.09 |
ENST00000528387.1
ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576
SRRM5
|
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr1_+_43855560 | 0.09 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr10_-_103815874 | 0.09 |
ENST00000370033.4
ENST00000311122.5 |
C10orf76
|
chromosome 10 open reading frame 76 |
| chr4_+_159593271 | 0.09 |
ENST00000512251.1
ENST00000511912.1 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr1_-_151300160 | 0.09 |
ENST00000368874.4
|
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr1_-_16539094 | 0.09 |
ENST00000270747.3
|
ARHGEF19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr17_-_4852332 | 0.09 |
ENST00000572383.1
|
PFN1
|
profilin 1 |
| chr20_+_44486246 | 0.09 |
ENST00000255152.2
ENST00000454862.2 |
ZSWIM3
|
zinc finger, SWIM-type containing 3 |
| chr16_-_69448 | 0.09 |
ENST00000326592.9
|
WASH4P
|
WAS protein family homolog 4 pseudogene |
| chr17_-_2615031 | 0.09 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr15_+_42120283 | 0.09 |
ENST00000542534.2
ENST00000397299.4 ENST00000408047.1 ENST00000431823.1 ENST00000382448.4 ENST00000342159.4 |
PLA2G4B
JMJD7
JMJD7-PLA2G4B
|
phospholipase A2, group IVB (cytosolic) jumonji domain containing 7 JMJD7-PLA2G4B readthrough |
| chr12_+_57916466 | 0.09 |
ENST00000355673.3
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr2_-_208031542 | 0.09 |
ENST00000423015.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr19_-_14201776 | 0.09 |
ENST00000269724.5
|
SAMD1
|
sterile alpha motif domain containing 1 |
| chr16_+_2255710 | 0.09 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chrX_+_51928002 | 0.09 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chr5_+_1801503 | 0.09 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr19_+_535835 | 0.09 |
ENST00000607527.1
ENST00000606065.1 |
CDC34
|
cell division cycle 34 |
| chr1_-_151299842 | 0.09 |
ENST00000438243.2
ENST00000489223.2 ENST00000368873.1 ENST00000430800.1 ENST00000368872.1 |
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr12_-_56727487 | 0.09 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr1_-_169555709 | 0.09 |
ENST00000546081.1
|
F5
|
coagulation factor V (proaccelerin, labile factor) |
| chr17_-_79817091 | 0.09 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr1_+_202317855 | 0.09 |
ENST00000356764.2
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr19_+_45394477 | 0.09 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr14_-_77787198 | 0.08 |
ENST00000261534.4
|
POMT2
|
protein-O-mannosyltransferase 2 |
| chr7_-_72936608 | 0.08 |
ENST00000404251.1
|
BAZ1B
|
bromodomain adjacent to zinc finger domain, 1B |
| chr8_+_145582633 | 0.08 |
ENST00000540505.1
|
SLC52A2
|
solute carrier family 52 (riboflavin transporter), member 2 |
| chr9_-_139137648 | 0.08 |
ENST00000358701.5
|
QSOX2
|
quiescin Q6 sulfhydryl oxidase 2 |
| chr2_+_220436917 | 0.08 |
ENST00000243786.2
|
INHA
|
inhibin, alpha |
| chr14_+_23352374 | 0.08 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr2_-_198364552 | 0.08 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr1_-_6420737 | 0.08 |
ENST00000541130.1
ENST00000377845.3 |
ACOT7
|
acyl-CoA thioesterase 7 |
| chr7_-_1067968 | 0.08 |
ENST00000412051.1
|
C7orf50
|
chromosome 7 open reading frame 50 |
| chr11_+_57559005 | 0.08 |
ENST00000534647.1
|
CTNND1
|
catenin (cadherin-associated protein), delta 1 |
| chr7_-_642261 | 0.08 |
ENST00000400758.2
|
PRKAR1B
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr1_+_44679159 | 0.08 |
ENST00000315913.5
ENST00000372289.2 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr5_-_42812143 | 0.08 |
ENST00000514985.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr17_-_80023659 | 0.08 |
ENST00000578907.1
ENST00000577907.1 ENST00000578176.1 ENST00000582529.1 |
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr8_-_73793975 | 0.08 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr11_-_61659006 | 0.08 |
ENST00000278829.2
|
FADS3
|
fatty acid desaturase 3 |
| chr3_+_14989186 | 0.08 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr1_-_151300126 | 0.08 |
ENST00000368875.2
ENST00000529142.1 |
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr20_+_30946106 | 0.08 |
ENST00000375687.4
ENST00000542461.1 |
ASXL1
|
additional sex combs like 1 (Drosophila) |
| chr20_+_3713314 | 0.08 |
ENST00000254963.2
ENST00000542646.1 ENST00000399701.1 |
HSPA12B
|
heat shock 70kD protein 12B |
| chr6_-_33281979 | 0.08 |
ENST00000426633.2
ENST00000467025.1 |
TAPBP
|
TAP binding protein (tapasin) |
| chr8_-_71316021 | 0.08 |
ENST00000452400.2
|
NCOA2
|
nuclear receptor coactivator 2 |
| chr17_+_7123207 | 0.08 |
ENST00000584103.1
ENST00000579886.2 |
ACADVL
|
acyl-CoA dehydrogenase, very long chain |
| chr17_+_27369918 | 0.08 |
ENST00000323372.4
|
PIPOX
|
pipecolic acid oxidase |
| chr17_-_46716647 | 0.08 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr20_+_37590942 | 0.08 |
ENST00000373325.2
ENST00000252011.3 ENST00000373323.4 |
DHX35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr4_-_109541539 | 0.08 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr2_+_27282134 | 0.08 |
ENST00000441931.1
|
AGBL5
|
ATP/GTP binding protein-like 5 |
| chr12_+_113659234 | 0.08 |
ENST00000551096.1
ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1
|
two pore segment channel 1 |
| chr12_-_7079805 | 0.08 |
ENST00000536316.2
ENST00000542912.1 ENST00000440277.1 ENST00000545167.1 ENST00000546111.1 ENST00000399433.2 ENST00000535923.1 |
PHB2
|
prohibitin 2 |
| chr20_+_32399093 | 0.08 |
ENST00000217402.2
|
CHMP4B
|
charged multivesicular body protein 4B |
| chr16_+_30075783 | 0.08 |
ENST00000412304.2
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr15_+_101402041 | 0.08 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr15_-_41120896 | 0.07 |
ENST00000299174.5
ENST00000427255.2 |
PPP1R14D
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr12_+_132413798 | 0.07 |
ENST00000440818.2
ENST00000542167.2 ENST00000538037.1 ENST00000456665.2 |
PUS1
|
pseudouridylate synthase 1 |
| chr16_-_4401258 | 0.07 |
ENST00000577031.1
|
PAM16
|
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr12_+_10365082 | 0.07 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr19_+_11546093 | 0.07 |
ENST00000591462.1
|
PRKCSH
|
protein kinase C substrate 80K-H |
| chr6_-_33282024 | 0.07 |
ENST00000475304.1
ENST00000489157.1 |
TAPBP
|
TAP binding protein (tapasin) |
| chr14_-_69350920 | 0.07 |
ENST00000553290.1
|
ACTN1
|
actinin, alpha 1 |
| chr6_-_30524951 | 0.07 |
ENST00000376621.3
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr19_+_1205740 | 0.07 |
ENST00000326873.7
|
STK11
|
serine/threonine kinase 11 |
| chr14_-_25519095 | 0.07 |
ENST00000419632.2
ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6
|
syntaxin binding protein 6 (amisyn) |
| chr22_-_45609315 | 0.07 |
ENST00000443310.3
ENST00000424508.1 |
KIAA0930
|
KIAA0930 |
| chr5_+_133842243 | 0.07 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr19_+_50145328 | 0.07 |
ENST00000360565.3
|
SCAF1
|
SR-related CTD-associated factor 1 |
| chr20_+_39969519 | 0.07 |
ENST00000373257.3
|
LPIN3
|
lipin 3 |
| chr2_-_85108240 | 0.07 |
ENST00000409133.1
|
TRABD2A
|
TraB domain containing 2A |
| chr11_+_7595136 | 0.07 |
ENST00000529575.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr22_+_25348671 | 0.07 |
ENST00000406486.4
|
KIAA1671
|
KIAA1671 |
| chr5_-_136834263 | 0.07 |
ENST00000505690.1
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr6_-_31926629 | 0.07 |
ENST00000375425.5
ENST00000426722.1 ENST00000441998.1 ENST00000444811.2 ENST00000375429.3 |
NELFE
|
negative elongation factor complex member E |
| chr1_+_113217073 | 0.07 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr18_+_54318566 | 0.07 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr19_-_34012674 | 0.07 |
ENST00000436370.3
ENST00000397032.4 ENST00000244137.7 |
PEPD
|
peptidase D |
| chr6_+_30882108 | 0.07 |
ENST00000541562.1
ENST00000421263.1 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr20_+_43374421 | 0.07 |
ENST00000372861.3
|
KCNK15
|
potassium channel, subfamily K, member 15 |
| chr17_-_7531121 | 0.07 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr22_+_38453378 | 0.07 |
ENST00000437453.1
ENST00000356976.3 |
PICK1
|
protein interacting with PRKCA 1 |
| chr12_+_107168418 | 0.07 |
ENST00000392839.2
ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B
|
RIC8 guanine nucleotide exchange factor B |
| chr12_+_132413765 | 0.07 |
ENST00000376649.3
ENST00000322060.5 |
PUS1
|
pseudouridylate synthase 1 |
| chr15_+_91449971 | 0.07 |
ENST00000557865.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr16_+_4475806 | 0.06 |
ENST00000262375.6
ENST00000355296.4 ENST00000431375.2 ENST00000574895.1 |
DNAJA3
|
DnaJ (Hsp40) homolog, subfamily A, member 3 |
| chr17_-_1395954 | 0.06 |
ENST00000359786.5
|
MYO1C
|
myosin IC |
| chr12_-_56367101 | 0.06 |
ENST00000549233.2
|
PMEL
|
premelanosome protein |
| chr1_-_29508321 | 0.06 |
ENST00000546138.1
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr22_+_21321447 | 0.06 |
ENST00000434714.1
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr3_-_49933506 | 0.06 |
ENST00000440292.1
|
MST1R
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr19_+_10222189 | 0.06 |
ENST00000321826.4
|
P2RY11
|
purinergic receptor P2Y, G-protein coupled, 11 |
| chrX_-_1511617 | 0.06 |
ENST00000381401.5
|
SLC25A6
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr10_+_106113515 | 0.06 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr17_-_2614927 | 0.06 |
ENST00000435359.1
|
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr17_-_15165825 | 0.06 |
ENST00000426385.3
|
PMP22
|
peripheral myelin protein 22 |
| chr1_-_154946825 | 0.06 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr17_-_7155775 | 0.06 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr22_+_46663895 | 0.06 |
ENST00000421359.1
|
TTC38
|
tetratricopeptide repeat domain 38 |
| chr1_+_12079517 | 0.06 |
ENST00000235332.4
ENST00000436478.2 |
MIIP
|
migration and invasion inhibitory protein |
| chr9_+_133986782 | 0.06 |
ENST00000372301.2
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr6_+_30594619 | 0.06 |
ENST00000318999.7
ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1
|
alpha tubulin acetyltransferase 1 |
| chr2_+_227771404 | 0.06 |
ENST00000409053.1
|
RHBDD1
|
rhomboid domain containing 1 |
| chr3_-_150264272 | 0.06 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr1_+_165864821 | 0.06 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr1_+_36621174 | 0.06 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr12_+_49717081 | 0.06 |
ENST00000547807.1
ENST00000551567.1 |
TROAP
|
trophinin associated protein |
| chr1_+_113217043 | 0.06 |
ENST00000413052.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr2_+_217498105 | 0.06 |
ENST00000233809.4
|
IGFBP2
|
insulin-like growth factor binding protein 2, 36kDa |
| chr3_-_47517302 | 0.06 |
ENST00000441517.2
ENST00000545718.1 |
SCAP
|
SREBF chaperone |
| chr5_+_170846640 | 0.06 |
ENST00000274625.5
|
FGF18
|
fibroblast growth factor 18 |
| chr17_-_4852243 | 0.06 |
ENST00000225655.5
|
PFN1
|
profilin 1 |
| chr1_-_156722195 | 0.06 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr19_+_10196981 | 0.06 |
ENST00000591813.1
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr7_-_102232891 | 0.06 |
ENST00000514917.2
|
RP11-514P8.7
|
RP11-514P8.7 |
| chr12_+_10365404 | 0.06 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr9_+_116263778 | 0.06 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr18_+_54318616 | 0.06 |
ENST00000254442.3
|
WDR7
|
WD repeat domain 7 |
| chr17_+_37821593 | 0.06 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chrX_+_53111541 | 0.06 |
ENST00000375442.4
ENST00000579390.1 |
TSPYL2
|
TSPY-like 2 |
| chr22_-_51017084 | 0.06 |
ENST00000360719.2
ENST00000457250.1 ENST00000440709.1 |
CPT1B
|
carnitine palmitoyltransferase 1B (muscle) |
| chr22_+_37415676 | 0.06 |
ENST00000401419.3
|
MPST
|
mercaptopyruvate sulfurtransferase |
| chr14_-_24658053 | 0.06 |
ENST00000354464.6
|
IPO4
|
importin 4 |
| chr1_+_113217345 | 0.06 |
ENST00000357443.2
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
Network of associatons between targets according to the STRING database.
First level regulatory network of RXRA_NR2F6_NR2C2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.1 | 0.2 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.2 | GO:0050823 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 | 0.2 | GO:0009440 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.4 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.1 | GO:0052510 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.1 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:1904808 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.0 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.0 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.0 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.0 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.0 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.0 | 0.1 | GO:0019474 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.0 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.0 | GO:1903939 | regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.0 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.0 | GO:0060769 | positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 0.0 | 0.0 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.1 | 0.2 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.0 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.0 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0005011 | macrophage colony-stimulating factor receptor activity(GO:0005011) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.0 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |