Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
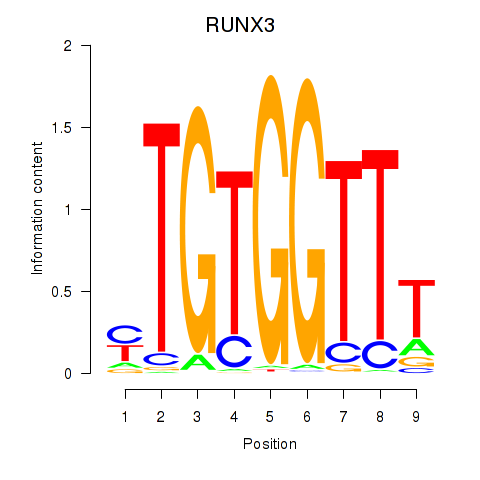
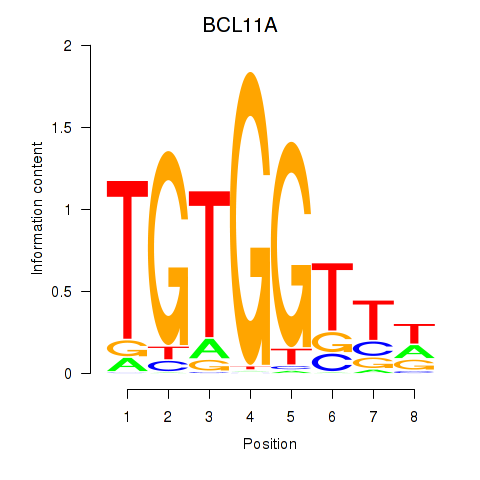
Results for RUNX3_BCL11A
Z-value: 0.83
Transcription factors associated with RUNX3_BCL11A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RUNX3
|
ENSG00000020633.14 | RUNX family transcription factor 3 |
|
BCL11A
|
ENSG00000119866.16 | BAF chromatin remodeling complex subunit BCL11A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BCL11A | hg19_v2_chr2_-_60780702_60780742 | 0.89 | 1.9e-02 | Click! |
| RUNX3 | hg19_v2_chr1_-_25256368_25256476 | -0.14 | 7.9e-01 | Click! |
Activity profile of RUNX3_BCL11A motif
Sorted Z-values of RUNX3_BCL11A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_73780852 | 0.99 |
ENST00000589666.1
|
UNK
|
unkempt family zinc finger |
| chr11_-_1606513 | 0.92 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr1_+_239882842 | 0.75 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr1_+_156211753 | 0.54 |
ENST00000368272.4
|
BGLAP
|
bone gamma-carboxyglutamate (gla) protein |
| chr12_-_58145889 | 0.53 |
ENST00000547853.1
|
CDK4
|
cyclin-dependent kinase 4 |
| chr7_-_84121858 | 0.50 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr1_-_168513229 | 0.47 |
ENST00000367819.2
|
XCL2
|
chemokine (C motif) ligand 2 |
| chr6_+_159071015 | 0.38 |
ENST00000360448.3
|
SYTL3
|
synaptotagmin-like 3 |
| chr17_+_75316336 | 0.36 |
ENST00000591934.1
|
SEPT9
|
septin 9 |
| chr1_-_206945830 | 0.36 |
ENST00000423557.1
|
IL10
|
interleukin 10 |
| chr17_-_67057047 | 0.35 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr14_+_64680854 | 0.34 |
ENST00000458046.2
|
SYNE2
|
spectrin repeat containing, nuclear envelope 2 |
| chrX_-_19688475 | 0.34 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr17_-_2169425 | 0.33 |
ENST00000570606.1
ENST00000354901.4 |
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr7_+_45067265 | 0.33 |
ENST00000474617.1
|
CCM2
|
cerebral cavernous malformation 2 |
| chr17_-_36884451 | 0.33 |
ENST00000595377.1
|
AC006449.1
|
NS5ATP13TP1; Uncharacterized protein |
| chr1_+_110453462 | 0.32 |
ENST00000488198.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr6_+_45296048 | 0.32 |
ENST00000465038.2
ENST00000352853.5 ENST00000541979.1 ENST00000371438.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr19_-_13900972 | 0.32 |
ENST00000397557.1
|
AC008686.1
|
Uncharacterized protein |
| chr3_+_187957646 | 0.31 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_+_62475130 | 0.31 |
ENST00000294117.5
|
GNG3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr5_+_131993856 | 0.30 |
ENST00000304506.3
|
IL13
|
interleukin 13 |
| chr1_+_23695680 | 0.28 |
ENST00000454117.1
ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213
|
chromosome 1 open reading frame 213 |
| chr3_+_49977894 | 0.27 |
ENST00000433811.1
|
RBM6
|
RNA binding motif protein 6 |
| chrX_+_37639264 | 0.27 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chrX_-_128788914 | 0.27 |
ENST00000429967.1
ENST00000307484.6 |
APLN
|
apelin |
| chr11_-_65548265 | 0.27 |
ENST00000532090.2
|
AP5B1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr6_+_31514622 | 0.26 |
ENST00000376146.4
|
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr1_-_94147385 | 0.26 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr1_+_110453203 | 0.26 |
ENST00000357302.4
ENST00000344188.5 ENST00000329608.6 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr1_+_12524965 | 0.25 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_+_110453608 | 0.24 |
ENST00000369801.1
|
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr13_+_28527647 | 0.24 |
ENST00000567234.1
|
LINC00543
|
long intergenic non-protein coding RNA 543 |
| chr19_-_51875894 | 0.24 |
ENST00000600427.1
ENST00000595217.1 ENST00000221978.5 |
NKG7
|
natural killer cell group 7 sequence |
| chr1_+_221051699 | 0.23 |
ENST00000366903.6
|
HLX
|
H2.0-like homeobox |
| chr15_+_86098670 | 0.23 |
ENST00000558811.1
|
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr4_-_84035905 | 0.23 |
ENST00000311507.4
|
PLAC8
|
placenta-specific 8 |
| chr1_+_44889697 | 0.23 |
ENST00000443020.2
|
RNF220
|
ring finger protein 220 |
| chr15_+_67390920 | 0.23 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chr17_+_40119801 | 0.23 |
ENST00000585452.1
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr6_+_15401075 | 0.23 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr20_+_43992094 | 0.23 |
ENST00000453003.1
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr2_+_74741569 | 0.22 |
ENST00000233638.7
|
TLX2
|
T-cell leukemia homeobox 2 |
| chr14_+_23352374 | 0.21 |
ENST00000267396.4
ENST00000536884.1 |
REM2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chrX_+_37639302 | 0.21 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr1_+_26872324 | 0.21 |
ENST00000531382.1
|
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chrX_+_48542168 | 0.21 |
ENST00000376701.4
|
WAS
|
Wiskott-Aldrich syndrome |
| chr8_-_11325047 | 0.20 |
ENST00000531804.1
|
FAM167A
|
family with sequence similarity 167, member A |
| chr5_+_176074388 | 0.19 |
ENST00000310032.8
ENST00000405525.2 |
TSPAN17
|
tetraspanin 17 |
| chr10_+_17270214 | 0.19 |
ENST00000544301.1
|
VIM
|
vimentin |
| chr12_-_4754339 | 0.19 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr3_+_136676707 | 0.19 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr3_+_141105235 | 0.18 |
ENST00000503809.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr19_+_36203830 | 0.18 |
ENST00000262630.3
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr8_+_145162629 | 0.18 |
ENST00000323662.8
|
KIAA1875
|
KIAA1875 |
| chr12_-_96793142 | 0.18 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr1_+_179051160 | 0.18 |
ENST00000367625.4
ENST00000352445.6 |
TOR3A
|
torsin family 3, member A |
| chr1_+_202385953 | 0.18 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr19_+_10947251 | 0.17 |
ENST00000592854.1
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr17_+_79935418 | 0.17 |
ENST00000306729.7
ENST00000306739.4 |
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr8_-_102803163 | 0.17 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr5_+_132149017 | 0.17 |
ENST00000378693.2
|
SOWAHA
|
sosondowah ankyrin repeat domain family member A |
| chr6_+_31950150 | 0.17 |
ENST00000537134.1
|
C4A
|
complement component 4A (Rodgers blood group) |
| chr1_+_110453514 | 0.17 |
ENST00000369802.3
ENST00000420111.2 |
CSF1
|
colony stimulating factor 1 (macrophage) |
| chr1_+_39876151 | 0.16 |
ENST00000530275.1
|
KIAA0754
|
KIAA0754 |
| chr1_-_207095324 | 0.16 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr10_+_17272608 | 0.16 |
ENST00000421459.2
|
VIM
|
vimentin |
| chr11_+_7559485 | 0.15 |
ENST00000527790.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr17_+_73455788 | 0.15 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr6_+_31515337 | 0.15 |
ENST00000376148.4
ENST00000376145.4 |
NFKBIL1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr15_-_37392724 | 0.15 |
ENST00000424352.2
|
MEIS2
|
Meis homeobox 2 |
| chr10_-_29811456 | 0.15 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr7_-_73184588 | 0.15 |
ENST00000395145.2
|
CLDN3
|
claudin 3 |
| chr18_-_5238525 | 0.15 |
ENST00000581170.1
ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5
LINC00526
|
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr19_-_10946871 | 0.15 |
ENST00000589638.1
|
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr5_+_67535647 | 0.15 |
ENST00000520675.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr14_-_23564320 | 0.14 |
ENST00000605057.1
|
ACIN1
|
apoptotic chromatin condensation inducer 1 |
| chr16_+_2570431 | 0.14 |
ENST00000563556.1
|
AMDHD2
|
amidohydrolase domain containing 2 |
| chr22_-_30642728 | 0.14 |
ENST00000403987.3
|
LIF
|
leukemia inhibitory factor |
| chr2_+_30455016 | 0.14 |
ENST00000401506.1
ENST00000407930.2 |
LBH
|
limb bud and heart development |
| chrX_-_40036520 | 0.14 |
ENST00000406200.2
ENST00000378455.4 ENST00000342274.4 |
BCOR
|
BCL6 corepressor |
| chr12_-_71031185 | 0.14 |
ENST00000548122.1
ENST00000551525.1 ENST00000550358.1 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr12_-_120662499 | 0.14 |
ENST00000552550.1
|
PXN
|
paxillin |
| chr3_+_50126341 | 0.14 |
ENST00000347869.3
ENST00000469838.1 ENST00000404526.2 ENST00000441305.1 |
RBM5
|
RNA binding motif protein 5 |
| chr1_+_198608146 | 0.13 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr15_-_37393406 | 0.13 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr5_-_94417186 | 0.13 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr19_-_6415695 | 0.13 |
ENST00000594496.1
ENST00000594745.1 ENST00000600480.1 |
KHSRP
|
KH-type splicing regulatory protein |
| chr17_+_79935464 | 0.13 |
ENST00000581647.1
ENST00000580534.1 ENST00000579684.1 |
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr4_-_6474173 | 0.13 |
ENST00000382599.4
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr5_+_35856951 | 0.13 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr19_+_13135439 | 0.13 |
ENST00000586873.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_+_33359473 | 0.13 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr17_+_7461849 | 0.13 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_-_128457446 | 0.13 |
ENST00000392668.4
|
ETS1
|
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr2_+_97203082 | 0.13 |
ENST00000454558.2
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr5_+_148206156 | 0.13 |
ENST00000305988.4
|
ADRB2
|
adrenoceptor beta 2, surface |
| chr1_+_145727681 | 0.13 |
ENST00000417171.1
ENST00000451928.2 |
PDZK1
|
PDZ domain containing 1 |
| chr11_+_86502085 | 0.13 |
ENST00000527521.1
|
PRSS23
|
protease, serine, 23 |
| chr17_-_4545170 | 0.13 |
ENST00000576394.1
ENST00000574640.1 |
ALOX15
|
arachidonate 15-lipoxygenase |
| chr16_-_89785777 | 0.13 |
ENST00000561976.1
|
VPS9D1
|
VPS9 domain containing 1 |
| chr17_+_40996590 | 0.13 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr4_-_152147579 | 0.12 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr2_-_119605253 | 0.12 |
ENST00000295206.6
|
EN1
|
engrailed homeobox 1 |
| chr11_-_62457371 | 0.12 |
ENST00000317449.4
|
LRRN4CL
|
LRRN4 C-terminal like |
| chr22_+_47016277 | 0.12 |
ENST00000406902.1
|
GRAMD4
|
GRAM domain containing 4 |
| chr7_+_139025875 | 0.12 |
ENST00000297534.6
|
C7orf55
|
chromosome 7 open reading frame 55 |
| chr16_+_2802623 | 0.12 |
ENST00000576924.1
ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr16_+_84801852 | 0.12 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr6_-_34524093 | 0.12 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr6_+_158733692 | 0.12 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr12_+_57916584 | 0.12 |
ENST00000546632.1
ENST00000549623.1 ENST00000431731.2 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr5_+_133451254 | 0.12 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr15_+_67418047 | 0.12 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr9_-_138853156 | 0.12 |
ENST00000371756.3
|
UBAC1
|
UBA domain containing 1 |
| chr6_+_30029008 | 0.12 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr19_-_50868882 | 0.12 |
ENST00000598915.1
|
NAPSA
|
napsin A aspartic peptidase |
| chr19_+_41257084 | 0.12 |
ENST00000601393.1
|
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr6_-_112081113 | 0.12 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr10_+_112404132 | 0.12 |
ENST00000369519.3
|
RBM20
|
RNA binding motif protein 20 |
| chr12_-_49245936 | 0.12 |
ENST00000308025.3
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr20_+_36405665 | 0.12 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr2_+_97202480 | 0.12 |
ENST00000357485.3
|
ARID5A
|
AT rich interactive domain 5A (MRF1-like) |
| chr5_+_176074455 | 0.12 |
ENST00000298564.10
ENST00000508164.1 ENST00000504168.1 ENST00000503045.1 ENST00000515708.1 ENST00000507471.1 |
TSPAN17
|
tetraspanin 17 |
| chr17_-_61776522 | 0.11 |
ENST00000582055.1
|
LIMD2
|
LIM domain containing 2 |
| chr11_+_36397528 | 0.11 |
ENST00000311599.5
ENST00000378867.3 |
PRR5L
|
proline rich 5 like |
| chr9_-_117160738 | 0.11 |
ENST00000448674.1
|
RP11-9M16.2
|
RP11-9M16.2 |
| chr19_-_49496557 | 0.11 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr1_+_179050512 | 0.11 |
ENST00000367627.3
|
TOR3A
|
torsin family 3, member A |
| chr6_+_157099036 | 0.11 |
ENST00000350026.5
ENST00000346085.5 ENST00000367148.1 ENST00000275248.4 |
ARID1B
|
AT rich interactive domain 1B (SWI1-like) |
| chr16_-_68482440 | 0.11 |
ENST00000219334.5
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
| chr22_-_30642782 | 0.11 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chr17_-_34625719 | 0.11 |
ENST00000422211.2
ENST00000542124.1 |
CCL3L1
|
chemokine (C-C motif) ligand 3-like 1 |
| chr1_-_157014865 | 0.11 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr4_-_57976544 | 0.11 |
ENST00000295666.4
ENST00000537922.1 |
IGFBP7
|
insulin-like growth factor binding protein 7 |
| chr3_+_189507523 | 0.11 |
ENST00000437221.1
ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63
|
tumor protein p63 |
| chr12_-_54121212 | 0.11 |
ENST00000548263.1
ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr9_-_123342415 | 0.11 |
ENST00000349780.4
ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr1_+_44679370 | 0.11 |
ENST00000372290.4
|
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr19_+_7580103 | 0.11 |
ENST00000596712.1
|
ZNF358
|
zinc finger protein 358 |
| chr5_-_94417339 | 0.11 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr6_+_144904334 | 0.11 |
ENST00000367526.4
|
UTRN
|
utrophin |
| chr3_-_196242233 | 0.11 |
ENST00000397537.2
|
SMCO1
|
single-pass membrane protein with coiled-coil domains 1 |
| chr17_-_61777090 | 0.11 |
ENST00000578061.1
|
LIMD2
|
LIM domain containing 2 |
| chr3_+_111260980 | 0.11 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr9_+_131799213 | 0.11 |
ENST00000358369.4
ENST00000406926.2 ENST00000277475.5 ENST00000450073.1 |
FAM73B
|
family with sequence similarity 73, member B |
| chr1_+_9299895 | 0.11 |
ENST00000602477.1
|
H6PD
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr12_-_120805872 | 0.11 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr7_-_84122033 | 0.10 |
ENST00000424555.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr3_-_131756559 | 0.10 |
ENST00000505957.1
|
CPNE4
|
copine IV |
| chr3_+_9944303 | 0.10 |
ENST00000421412.1
ENST00000295980.3 |
IL17RE
|
interleukin 17 receptor E |
| chr5_+_137722255 | 0.10 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr20_+_19870167 | 0.10 |
ENST00000440354.2
|
RIN2
|
Ras and Rab interactor 2 |
| chr12_+_57916466 | 0.10 |
ENST00000355673.3
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr17_-_46692287 | 0.10 |
ENST00000239144.4
|
HOXB8
|
homeobox B8 |
| chr5_-_94417314 | 0.10 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr4_+_71600063 | 0.10 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr19_-_40786733 | 0.10 |
ENST00000486368.2
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr13_-_38172863 | 0.10 |
ENST00000541481.1
ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN
|
periostin, osteoblast specific factor |
| chr1_-_150978953 | 0.10 |
ENST00000493834.2
|
FAM63A
|
family with sequence similarity 63, member A |
| chr3_+_189507460 | 0.10 |
ENST00000434928.1
|
TP63
|
tumor protein p63 |
| chr12_+_25205446 | 0.10 |
ENST00000557489.1
ENST00000354454.3 ENST00000536173.1 |
LRMP
|
lymphoid-restricted membrane protein |
| chr1_-_168464875 | 0.10 |
ENST00000422253.1
|
RP5-968D22.3
|
RP5-968D22.3 |
| chr19_+_36208877 | 0.09 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr1_+_155290677 | 0.09 |
ENST00000368354.3
ENST00000368352.5 |
RUSC1
|
RUN and SH3 domain containing 1 |
| chr3_-_99569821 | 0.09 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr17_+_19281034 | 0.09 |
ENST00000308406.5
ENST00000299612.7 |
MAPK7
|
mitogen-activated protein kinase 7 |
| chr5_-_138780159 | 0.09 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr6_+_42847649 | 0.09 |
ENST00000424341.2
ENST00000602561.1 |
RPL7L1
|
ribosomal protein L7-like 1 |
| chr17_+_76037081 | 0.09 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr12_-_54121261 | 0.09 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr14_+_85996471 | 0.09 |
ENST00000330753.4
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr1_-_12677714 | 0.09 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr12_-_58027002 | 0.09 |
ENST00000449184.3
|
B4GALNT1
|
beta-1,4-N-acetyl-galactosaminyl transferase 1 |
| chrX_+_155227371 | 0.09 |
ENST00000369423.2
ENST00000540897.1 |
IL9R
|
interleukin 9 receptor |
| chr19_+_49496705 | 0.09 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr5_-_94417562 | 0.09 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr17_+_75315534 | 0.09 |
ENST00000590294.1
ENST00000329047.8 |
SEPT9
|
septin 9 |
| chr1_-_98511756 | 0.09 |
ENST00000602984.1
ENST00000602852.1 |
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr1_-_25291475 | 0.09 |
ENST00000338888.3
ENST00000399916.1 |
RUNX3
|
runt-related transcription factor 3 |
| chr1_-_161277210 | 0.09 |
ENST00000491222.2
|
MPZ
|
myelin protein zero |
| chr17_+_75315654 | 0.09 |
ENST00000590595.1
|
SEPT9
|
septin 9 |
| chr5_+_131409476 | 0.09 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr7_+_18535321 | 0.08 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chr10_-_14372870 | 0.08 |
ENST00000357447.2
|
FRMD4A
|
FERM domain containing 4A |
| chr5_+_145316120 | 0.08 |
ENST00000359120.4
|
SH3RF2
|
SH3 domain containing ring finger 2 |
| chr6_+_143771934 | 0.08 |
ENST00000367592.1
|
PEX3
|
peroxisomal biogenesis factor 3 |
| chr12_-_48226897 | 0.08 |
ENST00000434070.1
|
HDAC7
|
histone deacetylase 7 |
| chr1_+_117544366 | 0.08 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chrX_+_155227246 | 0.08 |
ENST00000244174.5
ENST00000424344.3 |
IL9R
|
interleukin 9 receptor |
| chrX_-_47509887 | 0.08 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chr19_-_10946949 | 0.08 |
ENST00000214869.2
ENST00000591695.1 |
TMED1
|
transmembrane emp24 protein transport domain containing 1 |
| chr7_-_100844193 | 0.08 |
ENST00000440203.2
ENST00000379423.3 ENST00000223114.4 |
MOGAT3
|
monoacylglycerol O-acyltransferase 3 |
| chr3_+_51575596 | 0.08 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr15_-_78358465 | 0.08 |
ENST00000435468.1
|
TBC1D2B
|
TBC1 domain family, member 2B |
| chr3_-_196014520 | 0.08 |
ENST00000441879.1
ENST00000292823.2 ENST00000411591.1 ENST00000431016.1 ENST00000443555.1 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr11_-_67141090 | 0.08 |
ENST00000312438.7
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr11_-_67141640 | 0.08 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
| chr11_+_65292538 | 0.08 |
ENST00000270176.5
ENST00000525364.1 ENST00000420247.2 ENST00000533862.1 ENST00000279270.6 ENST00000524944.1 |
SCYL1
|
SCY1-like 1 (S. cerevisiae) |
| chr8_-_60031762 | 0.08 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
Network of associatons between targets according to the STRING database.
First level regulatory network of RUNX3_BCL11A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1902228 | mammary gland fat development(GO:0060611) positive regulation of macrophage colony-stimulating factor signaling pathway(GO:1902228) positive regulation of response to macrophage colony-stimulating factor(GO:1903971) positive regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903974) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) positive regulation of microglial cell migration(GO:1904141) |
| 0.2 | 0.6 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.2 | 0.8 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.1 | 0.4 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.5 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.1 | 0.5 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.3 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.4 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.5 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.2 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.3 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.1 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.2 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.3 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.2 | GO:0071104 | response to interleukin-9(GO:0071104) |
| 0.0 | 0.1 | GO:0070377 | regulation of ERK5 cascade(GO:0070376) negative regulation of ERK5 cascade(GO:0070377) |
| 0.0 | 0.2 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.4 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) NAD transport(GO:0043132) positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:2001178 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.6 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0048295 | positive regulation of isotype switching to IgE isotypes(GO:0048295) |
| 0.0 | 0.3 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.3 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:0010454 | negative regulation of cell fate commitment(GO:0010454) |
| 0.0 | 0.1 | GO:0097398 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.2 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.0 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 | 0.0 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 0.9 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.3 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.6 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) TOR complex(GO:0038201) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.7 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.9 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.3 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.3 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.1 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.0 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |