Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
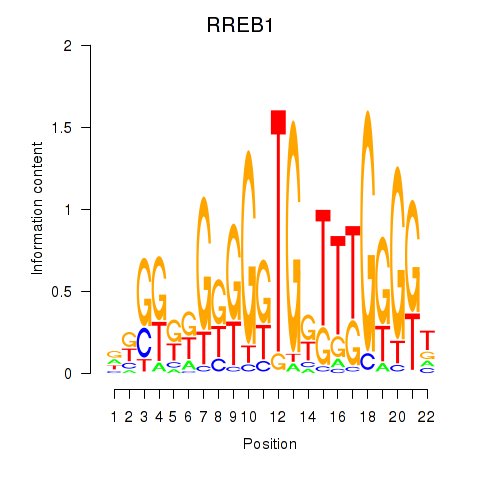
Results for RREB1
Z-value: 0.37
Transcription factors associated with RREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RREB1
|
ENSG00000124782.15 | ras responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RREB1 | hg19_v2_chr6_+_7107999_7108054 | 0.50 | 3.1e-01 | Click! |
Activity profile of RREB1 motif
Sorted Z-values of RREB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_20866424 | 0.22 |
ENST00000272224.3
|
GDF7
|
growth differentiation factor 7 |
| chr19_-_6720686 | 0.17 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr19_-_51071302 | 0.15 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr12_-_62653903 | 0.15 |
ENST00000552075.1
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr12_-_53297432 | 0.14 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr10_-_95360983 | 0.14 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr17_+_41177220 | 0.14 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr16_+_68771128 | 0.13 |
ENST00000261769.5
ENST00000422392.2 |
CDH1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr4_-_87374330 | 0.13 |
ENST00000511328.1
ENST00000503911.1 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr4_+_74735102 | 0.12 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr10_+_115438920 | 0.12 |
ENST00000429617.1
ENST00000369331.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr11_-_1033062 | 0.12 |
ENST00000525923.1
|
MUC6
|
mucin 6, oligomeric mucus/gel-forming |
| chr15_+_96897466 | 0.11 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chr4_-_103940791 | 0.10 |
ENST00000510559.1
ENST00000394789.3 ENST00000296422.7 |
SLC9B1
|
solute carrier family 9, subfamily B (NHA1, cation proton antiporter 1), member 1 |
| chr3_-_49203744 | 0.10 |
ENST00000321895.6
|
CCDC71
|
coiled-coil domain containing 71 |
| chr7_+_97840739 | 0.10 |
ENST00000609256.1
|
BHLHA15
|
basic helix-loop-helix family, member a15 |
| chr2_-_74645669 | 0.10 |
ENST00000518401.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr16_+_447209 | 0.09 |
ENST00000382940.4
ENST00000219479.2 |
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr8_+_38261880 | 0.09 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr19_-_18995029 | 0.09 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr11_+_1093318 | 0.09 |
ENST00000333592.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr17_-_73840614 | 0.09 |
ENST00000586108.1
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr17_+_15604513 | 0.08 |
ENST00000481540.1
|
ZNF286A
|
Homo sapiens zinc finger protein 286A (ZNF286A), transcript variant 6, mRNA. |
| chr16_+_447226 | 0.08 |
ENST00000433358.1
|
NME4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr19_+_36157715 | 0.08 |
ENST00000379013.2
ENST00000222275.2 |
UPK1A
|
uroplakin 1A |
| chr2_+_171571827 | 0.08 |
ENST00000375281.3
|
SP5
|
Sp5 transcription factor |
| chr16_+_57679859 | 0.08 |
ENST00000569494.1
ENST00000566169.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr10_+_13628933 | 0.08 |
ENST00000417658.1
ENST00000320054.4 |
PRPF18
|
pre-mRNA processing factor 18 |
| chr17_-_41132088 | 0.08 |
ENST00000591916.1
ENST00000451885.2 ENST00000454303.1 |
PTGES3L
PTGES3L-AARSD1
|
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr11_-_64703354 | 0.07 |
ENST00000532246.1
ENST00000279168.2 |
GPHA2
|
glycoprotein hormone alpha 2 |
| chr2_+_105471969 | 0.07 |
ENST00000361360.2
|
POU3F3
|
POU class 3 homeobox 3 |
| chr20_-_36793663 | 0.07 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr17_-_31620006 | 0.07 |
ENST00000225823.2
|
ASIC2
|
acid-sensing (proton-gated) ion channel 2 |
| chr1_+_61548374 | 0.07 |
ENST00000485903.2
ENST00000371185.2 ENST00000371184.2 |
NFIA
|
nuclear factor I/A |
| chr12_-_8088773 | 0.07 |
ENST00000544291.1
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr12_+_133033703 | 0.07 |
ENST00000542627.1
|
RP11-503G7.1
|
RP11-503G7.1 |
| chr3_+_192958914 | 0.07 |
ENST00000264735.2
ENST00000602513.1 |
HRASLS
|
HRAS-like suppressor |
| chr19_+_40973049 | 0.06 |
ENST00000598249.1
ENST00000338932.3 ENST00000344104.3 |
SPTBN4
|
spectrin, beta, non-erythrocytic 4 |
| chr12_+_54384370 | 0.06 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr19_-_30205963 | 0.06 |
ENST00000392278.2
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr22_+_38321840 | 0.06 |
ENST00000454685.1
|
MICALL1
|
MICAL-like 1 |
| chr18_-_45456693 | 0.06 |
ENST00000587421.1
|
SMAD2
|
SMAD family member 2 |
| chr22_-_37976082 | 0.06 |
ENST00000215886.4
|
LGALS2
|
lectin, galactoside-binding, soluble, 2 |
| chr14_-_105635090 | 0.06 |
ENST00000331782.3
ENST00000347004.2 |
JAG2
|
jagged 2 |
| chr4_-_100484825 | 0.06 |
ENST00000273962.3
ENST00000514547.1 ENST00000455368.2 |
TRMT10A
|
tRNA methyltransferase 10 homolog A (S. cerevisiae) |
| chr1_-_228604544 | 0.06 |
ENST00000457345.2
|
TRIM17
|
tripartite motif containing 17 |
| chr22_+_41968007 | 0.06 |
ENST00000460790.1
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr19_+_55795493 | 0.06 |
ENST00000309383.1
|
BRSK1
|
BR serine/threonine kinase 1 |
| chr8_-_22550691 | 0.06 |
ENST00000519492.1
|
EGR3
|
early growth response 3 |
| chr4_+_108746282 | 0.06 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr6_+_151646800 | 0.06 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr1_-_31712401 | 0.06 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr12_+_54447637 | 0.06 |
ENST00000609810.1
ENST00000430889.2 |
HOXC4
HOXC4
|
homeobox C4 Homeobox protein Hox-C4 |
| chr16_-_3285049 | 0.06 |
ENST00000575948.1
|
ZNF200
|
zinc finger protein 200 |
| chr11_+_66406088 | 0.06 |
ENST00000310092.7
ENST00000396053.4 ENST00000408993.2 |
RBM4
|
RNA binding motif protein 4 |
| chr7_-_65113280 | 0.06 |
ENST00000593865.1
|
AC104057.1
|
Protein LOC100996407 |
| chr4_-_6474173 | 0.06 |
ENST00000382599.4
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr11_+_46403303 | 0.06 |
ENST00000407067.1
ENST00000395565.1 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr16_+_57680043 | 0.06 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr2_+_85843252 | 0.05 |
ENST00000409025.1
ENST00000409470.1 ENST00000323701.6 ENST00000409766.3 |
USP39
|
ubiquitin specific peptidase 39 |
| chr17_-_79817091 | 0.05 |
ENST00000570907.1
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide |
| chr17_-_39216344 | 0.05 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr12_-_12715266 | 0.05 |
ENST00000228862.2
|
DUSP16
|
dual specificity phosphatase 16 |
| chr11_+_46403194 | 0.05 |
ENST00000395569.4
ENST00000395566.4 |
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr4_+_164265035 | 0.05 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr3_+_187461442 | 0.05 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr8_+_32406137 | 0.05 |
ENST00000521670.1
|
NRG1
|
neuregulin 1 |
| chr19_-_18654293 | 0.05 |
ENST00000597547.1
ENST00000222308.4 ENST00000544835.3 ENST00000610101.1 ENST00000597960.3 ENST00000608443.1 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr1_-_149889382 | 0.05 |
ENST00000369145.1
ENST00000369146.3 |
SV2A
|
synaptic vesicle glycoprotein 2A |
| chr15_-_70390213 | 0.05 |
ENST00000557997.1
ENST00000317509.8 ENST00000442299.2 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr12_-_49351148 | 0.05 |
ENST00000398092.4
ENST00000539611.1 |
RP11-302B13.5
ARF3
|
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr8_-_66754172 | 0.05 |
ENST00000401827.3
|
PDE7A
|
phosphodiesterase 7A |
| chr17_-_58156273 | 0.05 |
ENST00000593228.1
ENST00000585976.1 ENST00000593097.1 ENST00000184956.6 |
HEATR6
|
HEAT repeat containing 6 |
| chr19_-_42759266 | 0.05 |
ENST00000594664.1
|
AC006486.9
|
Uncharacterized protein |
| chr19_-_49140609 | 0.05 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr12_+_54402790 | 0.05 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr1_-_113392399 | 0.05 |
ENST00000449572.2
ENST00000433505.1 |
RP11-426L16.8
|
RP11-426L16.8 |
| chr17_+_30348024 | 0.04 |
ENST00000327564.7
ENST00000584368.1 ENST00000394713.3 ENST00000341671.7 |
LRRC37B
|
leucine rich repeat containing 37B |
| chr16_+_30969055 | 0.04 |
ENST00000452917.1
|
SETD1A
|
SET domain containing 1A |
| chr10_-_121356518 | 0.04 |
ENST00000369092.4
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr11_-_17498348 | 0.04 |
ENST00000389817.3
ENST00000302539.4 |
ABCC8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr13_+_111972980 | 0.04 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chr1_+_154975110 | 0.04 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr13_-_99852916 | 0.04 |
ENST00000426037.2
ENST00000445737.2 |
UBAC2-AS1
|
UBAC2 antisense RNA 1 |
| chr14_-_71067360 | 0.04 |
ENST00000554963.1
ENST00000430055.2 ENST00000440435.2 ENST00000256379.5 |
MED6
|
mediator complex subunit 6 |
| chrX_-_151903184 | 0.04 |
ENST00000357916.4
ENST00000393869.3 |
MAGEA12
|
melanoma antigen family A, 12 |
| chr1_-_28559502 | 0.04 |
ENST00000263697.4
|
DNAJC8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr17_-_7082861 | 0.04 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chrX_+_53078465 | 0.04 |
ENST00000375466.2
|
GPR173
|
G protein-coupled receptor 173 |
| chr16_-_4588762 | 0.04 |
ENST00000562334.1
ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr4_-_77996032 | 0.04 |
ENST00000505609.1
|
CCNI
|
cyclin I |
| chr19_-_49140692 | 0.04 |
ENST00000222122.5
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr17_+_7482785 | 0.04 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr12_+_56415100 | 0.04 |
ENST00000547791.1
|
IKZF4
|
IKAROS family zinc finger 4 (Eos) |
| chr3_+_48507210 | 0.04 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr14_+_74353508 | 0.04 |
ENST00000324593.6
ENST00000557495.1 ENST00000556659.1 ENST00000557363.1 |
ZNF410
|
zinc finger protein 410 |
| chr12_-_8088871 | 0.04 |
ENST00000075120.7
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr6_+_149638876 | 0.04 |
ENST00000392282.1
|
TAB2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr3_+_48507621 | 0.04 |
ENST00000456089.1
|
TREX1
|
three prime repair exonuclease 1 |
| chr19_-_3029011 | 0.04 |
ENST00000590536.1
ENST00000587137.1 ENST00000455444.2 ENST00000262953.6 |
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr16_+_27325202 | 0.04 |
ENST00000395762.2
ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R
|
interleukin 4 receptor |
| chr22_+_46449674 | 0.04 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr19_-_18653781 | 0.04 |
ENST00000596558.2
ENST00000453489.2 |
FKBP8
|
FK506 binding protein 8, 38kDa |
| chr6_-_136847610 | 0.04 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr11_+_46402744 | 0.04 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr17_-_4642429 | 0.04 |
ENST00000573123.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr12_+_49212514 | 0.04 |
ENST00000301050.2
ENST00000548279.1 ENST00000547230.1 |
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr5_+_151151471 | 0.04 |
ENST00000394123.3
ENST00000543466.1 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr12_-_57644952 | 0.04 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr4_+_75230853 | 0.04 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr19_-_8675559 | 0.03 |
ENST00000597188.1
|
ADAMTS10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
| chr19_+_15619299 | 0.03 |
ENST00000269703.3
|
CYP4F22
|
cytochrome P450, family 4, subfamily F, polypeptide 22 |
| chr3_+_53195517 | 0.03 |
ENST00000487897.1
|
PRKCD
|
protein kinase C, delta |
| chr19_+_50354462 | 0.03 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr7_-_154794621 | 0.03 |
ENST00000419436.1
ENST00000397192.1 |
PAXIP1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr17_+_48611853 | 0.03 |
ENST00000507709.1
ENST00000515126.1 ENST00000507467.1 |
EPN3
|
epsin 3 |
| chr1_+_154975258 | 0.03 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr15_+_77713299 | 0.03 |
ENST00000559099.1
|
HMG20A
|
high mobility group 20A |
| chr1_-_23857698 | 0.03 |
ENST00000361729.2
|
E2F2
|
E2F transcription factor 2 |
| chr11_-_57089671 | 0.03 |
ENST00000532437.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr21_+_44073860 | 0.03 |
ENST00000335512.4
ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A
|
phosphodiesterase 9A |
| chrX_+_128674213 | 0.03 |
ENST00000371113.4
ENST00000357121.5 |
OCRL
|
oculocerebrorenal syndrome of Lowe |
| chr22_-_36357671 | 0.03 |
ENST00000408983.2
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr17_-_6459768 | 0.03 |
ENST00000421306.3
|
PITPNM3
|
PITPNM family member 3 |
| chr18_+_56531584 | 0.03 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr15_+_21145765 | 0.03 |
ENST00000553416.1
|
CT60
|
cancer/testis antigen 60 (non-protein coding) |
| chr9_+_132427883 | 0.03 |
ENST00000372469.4
|
PRRX2
|
paired related homeobox 2 |
| chr16_+_57679945 | 0.03 |
ENST00000568157.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr19_+_46009837 | 0.03 |
ENST00000589627.1
|
VASP
|
vasodilator-stimulated phosphoprotein |
| chr1_+_157963063 | 0.03 |
ENST00000360089.4
ENST00000368173.3 ENST00000392272.2 |
KIRREL
|
kin of IRRE like (Drosophila) |
| chr5_+_92228 | 0.03 |
ENST00000512035.1
|
CTD-2231H16.1
|
CTD-2231H16.1 |
| chr11_-_8739383 | 0.03 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr16_+_67143880 | 0.03 |
ENST00000219139.3
ENST00000566026.1 |
C16orf70
|
chromosome 16 open reading frame 70 |
| chr1_-_6240183 | 0.03 |
ENST00000262450.3
ENST00000378021.1 |
CHD5
|
chromodomain helicase DNA binding protein 5 |
| chr7_-_154794763 | 0.03 |
ENST00000404141.1
|
PAXIP1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr2_+_242577097 | 0.03 |
ENST00000419606.1
ENST00000474739.2 ENST00000396411.3 ENST00000425239.1 ENST00000400771.3 ENST00000430617.2 |
ATG4B
|
autophagy related 4B, cysteine peptidase |
| chr14_+_74353574 | 0.03 |
ENST00000442160.3
ENST00000555044.1 |
ZNF410
|
zinc finger protein 410 |
| chr17_+_7788104 | 0.03 |
ENST00000380358.4
|
CHD3
|
chromodomain helicase DNA binding protein 3 |
| chr17_-_7141490 | 0.03 |
ENST00000574236.1
ENST00000572789.1 |
PHF23
|
PHD finger protein 23 |
| chr8_+_144816303 | 0.03 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr14_+_74353320 | 0.03 |
ENST00000540593.1
ENST00000555730.1 |
ZNF410
|
zinc finger protein 410 |
| chr12_+_58003935 | 0.03 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr12_+_57916584 | 0.03 |
ENST00000546632.1
ENST00000549623.1 ENST00000431731.2 |
MBD6
|
methyl-CpG binding domain protein 6 |
| chr12_+_132413765 | 0.03 |
ENST00000376649.3
ENST00000322060.5 |
PUS1
|
pseudouridylate synthase 1 |
| chrX_-_153881842 | 0.03 |
ENST00000369585.3
ENST00000247306.4 |
CTAG2
|
cancer/testis antigen 2 |
| chr8_-_22550815 | 0.03 |
ENST00000317216.2
|
EGR3
|
early growth response 3 |
| chr20_+_36888551 | 0.03 |
ENST00000418004.1
ENST00000451435.1 |
BPI
|
bactericidal/permeability-increasing protein |
| chr20_+_62697564 | 0.03 |
ENST00000458442.1
|
TCEA2
|
transcription elongation factor A (SII), 2 |
| chr11_-_61197406 | 0.03 |
ENST00000541963.1
ENST00000477890.2 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr3_-_125094093 | 0.02 |
ENST00000484491.1
ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148
|
zinc finger protein 148 |
| chr22_-_51066521 | 0.02 |
ENST00000395621.3
ENST00000395619.3 ENST00000356098.5 ENST00000216124.5 ENST00000453344.2 ENST00000547307.1 ENST00000547805.1 |
ARSA
|
arylsulfatase A |
| chr18_-_21242774 | 0.02 |
ENST00000322980.9
|
ANKRD29
|
ankyrin repeat domain 29 |
| chr2_-_202508169 | 0.02 |
ENST00000409883.2
|
TMEM237
|
transmembrane protein 237 |
| chr1_+_220267429 | 0.02 |
ENST00000366922.1
ENST00000302637.5 |
IARS2
|
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr1_-_226111929 | 0.02 |
ENST00000343818.6
ENST00000432920.2 |
PYCR2
RP4-559A3.7
|
pyrroline-5-carboxylate reductase family, member 2 Uncharacterized protein |
| chr8_+_32405785 | 0.02 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr16_-_85146040 | 0.02 |
ENST00000539556.1
|
FAM92B
|
family with sequence similarity 92, member B |
| chr17_-_76732928 | 0.02 |
ENST00000589768.1
|
CYTH1
|
cytohesin 1 |
| chr2_+_219264762 | 0.02 |
ENST00000452977.1
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chrX_+_102631844 | 0.02 |
ENST00000372634.1
ENST00000299872.7 |
NGFRAP1
|
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr6_+_7107830 | 0.02 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr20_+_48599506 | 0.02 |
ENST00000244050.2
|
SNAI1
|
snail family zinc finger 1 |
| chr22_-_38484922 | 0.02 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr2_+_48757278 | 0.02 |
ENST00000404752.1
ENST00000406226.1 |
STON1
|
stonin 1 |
| chr2_-_101034070 | 0.02 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr11_-_129872672 | 0.02 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr21_-_34915123 | 0.02 |
ENST00000438059.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr11_+_61197508 | 0.02 |
ENST00000541135.1
ENST00000301761.2 |
RP11-286N22.8
SDHAF2
|
Uncharacterized protein succinate dehydrogenase complex assembly factor 2 |
| chr15_-_70390191 | 0.02 |
ENST00000559191.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr2_-_242576864 | 0.02 |
ENST00000407315.1
|
THAP4
|
THAP domain containing 4 |
| chr2_-_220108309 | 0.02 |
ENST00000409640.1
|
GLB1L
|
galactosidase, beta 1-like |
| chr16_-_11922665 | 0.02 |
ENST00000573319.1
ENST00000577041.1 ENST00000574028.1 ENST00000571259.1 ENST00000573037.1 ENST00000571158.1 |
BCAR4
|
breast cancer anti-estrogen resistance 4 (non-protein coding) |
| chr15_-_23692381 | 0.02 |
ENST00000567107.1
ENST00000345070.5 ENST00000312015.5 |
GOLGA6L2
|
golgin A6 family-like 2 |
| chr16_+_50727479 | 0.02 |
ENST00000531674.1
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr1_-_204380919 | 0.02 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr5_+_151151504 | 0.02 |
ENST00000356245.3
ENST00000507878.2 |
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr10_+_52751010 | 0.02 |
ENST00000373985.1
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr1_-_155658085 | 0.02 |
ENST00000311573.5
ENST00000438245.2 |
YY1AP1
|
YY1 associated protein 1 |
| chr17_+_58499066 | 0.02 |
ENST00000474834.1
|
C17orf64
|
chromosome 17 open reading frame 64 |
| chr15_+_63481668 | 0.02 |
ENST00000321437.4
ENST00000559006.1 ENST00000448330.2 |
RAB8B
|
RAB8B, member RAS oncogene family |
| chr19_-_3025614 | 0.02 |
ENST00000447365.2
|
TLE2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr7_+_101459263 | 0.02 |
ENST00000292538.4
ENST00000393824.3 ENST00000547394.2 ENST00000360264.3 ENST00000425244.2 |
CUX1
|
cut-like homeobox 1 |
| chr18_+_9334786 | 0.02 |
ENST00000581641.1
|
TWSG1
|
twisted gastrulation BMP signaling modulator 1 |
| chr11_-_75062829 | 0.02 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr3_+_133524459 | 0.02 |
ENST00000484684.1
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chr5_-_142065612 | 0.02 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr5_+_169758393 | 0.02 |
ENST00000521471.1
ENST00000518357.1 ENST00000436248.3 |
CTB-114C7.3
|
CTB-114C7.3 |
| chr7_-_128045984 | 0.02 |
ENST00000470772.1
ENST00000480861.1 ENST00000496200.1 |
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr9_+_79634571 | 0.02 |
ENST00000376708.1
|
FOXB2
|
forkhead box B2 |
| chr11_-_75062730 | 0.02 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr17_-_42143963 | 0.02 |
ENST00000585388.1
ENST00000293406.3 |
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr5_-_124080203 | 0.02 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr19_+_55999771 | 0.02 |
ENST00000594321.1
|
SSC5D
|
scavenger receptor cysteine rich domain containing (5 domains) |
| chrX_+_17755563 | 0.02 |
ENST00000380045.3
ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1
|
sex comb on midleg-like 1 (Drosophila) |
| chr8_+_28748099 | 0.02 |
ENST00000519047.1
|
HMBOX1
|
homeobox containing 1 |
| chr5_-_98262240 | 0.02 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr7_+_94537247 | 0.02 |
ENST00000422324.1
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A |
| chr17_+_48796905 | 0.02 |
ENST00000505658.1
ENST00000393227.2 ENST00000240304.1 ENST00000311571.3 ENST00000505619.1 ENST00000544170.1 ENST00000510984.1 |
LUC7L3
|
LUC7-like 3 (S. cerevisiae) |
| chr8_-_66753682 | 0.02 |
ENST00000396642.3
|
PDE7A
|
phosphodiesterase 7A |
| chr14_-_24912047 | 0.02 |
ENST00000553930.1
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RREB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.0 | 0.2 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.1 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0018262 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.1 | GO:0072240 | ascending thin limb development(GO:0072021) DCT cell differentiation(GO:0072069) metanephric ascending thin limb development(GO:0072218) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.1 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.0 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0032090 | Pyrin domain binding(GO:0032090) |