Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
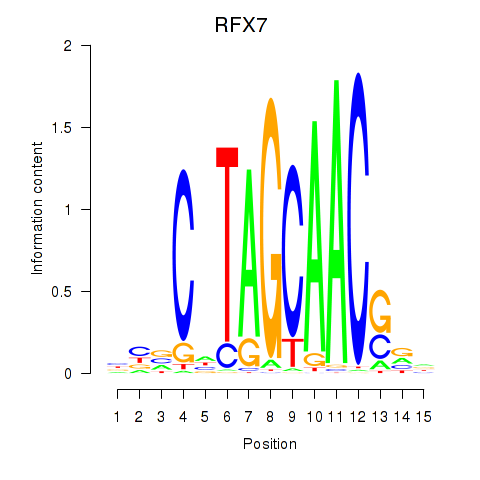
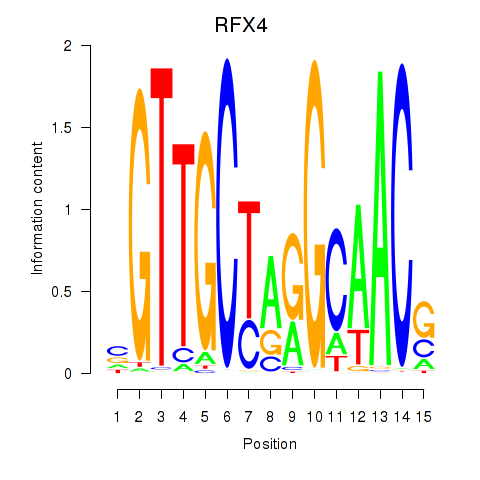
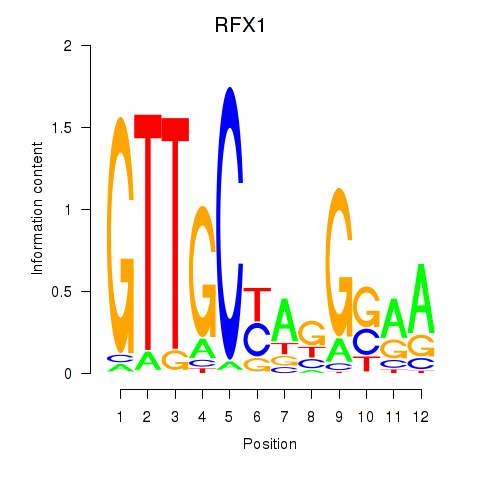
Results for RFX7_RFX4_RFX1
Z-value: 1.16
Transcription factors associated with RFX7_RFX4_RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX7
|
ENSG00000181827.10 | regulatory factor X7 |
|
RFX4
|
ENSG00000111783.8 | regulatory factor X4 |
|
RFX1
|
ENSG00000132005.4 | regulatory factor X1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX1 | hg19_v2_chr19_-_14117074_14117141 | -0.87 | 2.5e-02 | Click! |
| RFX4 | hg19_v2_chr12_+_107078474_107078533 | -0.68 | 1.4e-01 | Click! |
| RFX7 | hg19_v2_chr15_-_56535464_56535521 | 0.41 | 4.2e-01 | Click! |
Activity profile of RFX7_RFX4_RFX1 motif
Sorted Z-values of RFX7_RFX4_RFX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_76478386 | 1.50 |
ENST00000535020.2
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr10_+_25305524 | 1.33 |
ENST00000524413.1
ENST00000376356.4 |
THNSL1
|
threonine synthase-like 1 (S. cerevisiae) |
| chr12_-_76478417 | 1.23 |
ENST00000552342.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr12_-_76478446 | 1.07 |
ENST00000393263.3
ENST00000548044.1 ENST00000547704.1 ENST00000431879.3 ENST00000549596.1 ENST00000550934.1 ENST00000551600.1 ENST00000547479.1 ENST00000547773.1 ENST00000544816.1 ENST00000542344.1 ENST00000548273.1 |
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr1_-_85156090 | 1.05 |
ENST00000605755.1
ENST00000437941.2 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_-_85156417 | 1.02 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr15_-_49447771 | 0.98 |
ENST00000558843.1
ENST00000542928.1 ENST00000561248.1 |
COPS2
|
COP9 signalosome subunit 2 |
| chr1_-_85156216 | 0.97 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr2_+_172778952 | 0.95 |
ENST00000392584.1
ENST00000264108.4 |
HAT1
|
histone acetyltransferase 1 |
| chr17_+_2264983 | 0.92 |
ENST00000574650.1
|
SGSM2
|
small G protein signaling modulator 2 |
| chr1_+_45140400 | 0.90 |
ENST00000453711.1
|
C1orf228
|
chromosome 1 open reading frame 228 |
| chr12_-_76478686 | 0.87 |
ENST00000261182.8
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr22_-_23484246 | 0.86 |
ENST00000216036.4
|
RTDR1
|
rhabdoid tumor deletion region gene 1 |
| chr8_+_94767072 | 0.86 |
ENST00000452276.1
ENST00000453321.3 ENST00000498673.1 ENST00000518319.1 |
TMEM67
|
transmembrane protein 67 |
| chr1_-_86861660 | 0.84 |
ENST00000486215.1
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr9_+_79792269 | 0.84 |
ENST00000376634.4
ENST00000376636.3 ENST00000360280.3 |
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr4_-_156297949 | 0.81 |
ENST00000515654.1
|
MAP9
|
microtubule-associated protein 9 |
| chr1_-_74663825 | 0.75 |
ENST00000370911.3
ENST00000370909.2 ENST00000354431.4 |
LRRIQ3
|
leucine-rich repeats and IQ motif containing 3 |
| chr4_-_156298087 | 0.72 |
ENST00000311277.4
|
MAP9
|
microtubule-associated protein 9 |
| chr9_+_79792410 | 0.72 |
ENST00000357409.5
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr22_-_50970919 | 0.71 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr20_+_17550691 | 0.71 |
ENST00000474024.1
|
DSTN
|
destrin (actin depolymerizing factor) |
| chr19_+_2096868 | 0.70 |
ENST00000395296.1
ENST00000395301.3 |
IZUMO4
|
IZUMO family member 4 |
| chr13_+_103046954 | 0.70 |
ENST00000606448.1
|
FGF14-AS2
|
FGF14 antisense RNA 2 |
| chr15_-_49447835 | 0.70 |
ENST00000388901.5
ENST00000299259.6 |
COPS2
|
COP9 signalosome subunit 2 |
| chr10_+_15001430 | 0.69 |
ENST00000407572.1
|
MEIG1
|
meiosis/spermiogenesis associated 1 |
| chr1_+_74663994 | 0.69 |
ENST00000472069.1
|
FPGT
|
fucose-1-phosphate guanylyltransferase |
| chr3_-_52739670 | 0.64 |
ENST00000497953.1
|
GLT8D1
|
glycosyltransferase 8 domain containing 1 |
| chr5_+_68485433 | 0.63 |
ENST00000502689.1
|
CENPH
|
centromere protein H |
| chr8_+_59323823 | 0.63 |
ENST00000399598.2
|
UBXN2B
|
UBX domain protein 2B |
| chr22_-_32555275 | 0.62 |
ENST00000382097.3
|
C22orf42
|
chromosome 22 open reading frame 42 |
| chr12_-_107168696 | 0.62 |
ENST00000551505.1
|
RP11-144F15.1
|
Uncharacterized protein |
| chr8_-_93978216 | 0.61 |
ENST00000517751.1
ENST00000524107.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr15_+_71185148 | 0.61 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr2_-_27851843 | 0.60 |
ENST00000324364.3
|
CCDC121
|
coiled-coil domain containing 121 |
| chr16_-_85146040 | 0.57 |
ENST00000539556.1
|
FAM92B
|
family with sequence similarity 92, member B |
| chr15_-_71184724 | 0.54 |
ENST00000560604.1
|
THAP10
|
THAP domain containing 10 |
| chr2_+_54558004 | 0.54 |
ENST00000405749.1
ENST00000398634.2 ENST00000447328.1 |
C2orf73
|
chromosome 2 open reading frame 73 |
| chr21_-_43916433 | 0.54 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr22_-_50970566 | 0.53 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr5_-_133702761 | 0.51 |
ENST00000521118.1
ENST00000265334.4 ENST00000435211.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr12_+_100660909 | 0.51 |
ENST00000549687.1
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr15_-_55700216 | 0.50 |
ENST00000569205.1
|
CCPG1
|
cell cycle progression 1 |
| chr1_-_63988846 | 0.50 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr3_+_97483572 | 0.50 |
ENST00000335979.2
ENST00000394206.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr2_+_44001172 | 0.49 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr12_+_110562135 | 0.49 |
ENST00000361948.4
ENST00000552912.1 ENST00000242591.5 ENST00000546374.1 |
IFT81
|
intraflagellar transport 81 homolog (Chlamydomonas) |
| chr2_-_196933536 | 0.48 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chrX_+_23925918 | 0.48 |
ENST00000379211.3
|
CXorf58
|
chromosome X open reading frame 58 |
| chr20_+_17550489 | 0.48 |
ENST00000246069.7
|
DSTN
|
destrin (actin depolymerizing factor) |
| chr4_-_156298028 | 0.48 |
ENST00000433024.1
ENST00000379248.2 |
MAP9
|
microtubule-associated protein 9 |
| chr15_+_71184931 | 0.48 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr16_-_29757272 | 0.47 |
ENST00000329410.3
|
C16orf54
|
chromosome 16 open reading frame 54 |
| chr1_-_207226313 | 0.47 |
ENST00000367084.1
|
YOD1
|
YOD1 deubiquitinase |
| chr22_-_50970506 | 0.46 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr17_+_42733730 | 0.46 |
ENST00000359945.3
ENST00000425535.1 |
C17orf104
|
chromosome 17 open reading frame 104 |
| chr12_+_100660940 | 0.45 |
ENST00000548392.1
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr17_-_7108436 | 0.45 |
ENST00000493294.1
|
DLG4
|
discs, large homolog 4 (Drosophila) |
| chr3_+_132379154 | 0.45 |
ENST00000468022.1
ENST00000473651.1 ENST00000494238.2 |
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chr15_+_49913201 | 0.45 |
ENST00000329873.5
ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1
|
DTW domain containing 1 |
| chr6_+_142468361 | 0.44 |
ENST00000367630.4
|
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr17_+_42733803 | 0.44 |
ENST00000409122.2
|
C17orf104
|
chromosome 17 open reading frame 104 |
| chr1_-_159880159 | 0.44 |
ENST00000599780.1
|
AL590560.1
|
HCG1995379; Uncharacterized protein |
| chr3_-_48659193 | 0.44 |
ENST00000330862.3
|
TMEM89
|
transmembrane protein 89 |
| chr20_-_49575081 | 0.42 |
ENST00000371588.5
ENST00000371582.4 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr18_+_29671812 | 0.42 |
ENST00000261593.3
ENST00000578914.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chr9_+_26956371 | 0.41 |
ENST00000380062.5
ENST00000518614.1 |
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr8_-_93978357 | 0.41 |
ENST00000522925.1
ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr20_+_18447771 | 0.41 |
ENST00000377603.4
|
POLR3F
|
polymerase (RNA) III (DNA directed) polypeptide F, 39 kDa |
| chr15_-_55790515 | 0.41 |
ENST00000448430.2
ENST00000457155.2 |
DYX1C1
|
dyslexia susceptibility 1 candidate 1 |
| chr12_-_100660833 | 0.40 |
ENST00000551642.1
ENST00000416321.1 ENST00000550587.1 ENST00000549249.1 |
DEPDC4
|
DEP domain containing 4 |
| chr13_+_21141208 | 0.40 |
ENST00000351808.5
|
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr5_+_110074685 | 0.39 |
ENST00000355943.3
ENST00000447245.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr4_-_153700864 | 0.38 |
ENST00000304337.2
|
TIGD4
|
tigger transposable element derived 4 |
| chr2_-_63815628 | 0.38 |
ENST00000409562.3
|
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr2_-_47142884 | 0.38 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr4_+_26585686 | 0.38 |
ENST00000505206.1
ENST00000511789.1 |
TBC1D19
|
TBC1 domain family, member 19 |
| chr13_+_21141270 | 0.37 |
ENST00000319980.6
ENST00000537103.1 ENST00000389373.3 |
IFT88
|
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr4_+_6784401 | 0.37 |
ENST00000425103.1
ENST00000307659.5 |
KIAA0232
|
KIAA0232 |
| chr1_-_119683251 | 0.36 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr5_-_43313574 | 0.36 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr7_+_12726474 | 0.36 |
ENST00000396662.1
ENST00000356797.3 ENST00000396664.2 |
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr3_+_97483366 | 0.35 |
ENST00000463745.1
ENST00000462412.1 |
ARL6
|
ADP-ribosylation factor-like 6 |
| chr1_+_206808918 | 0.35 |
ENST00000367108.3
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr9_-_138391692 | 0.35 |
ENST00000429260.2
|
C9orf116
|
chromosome 9 open reading frame 116 |
| chr4_+_6784358 | 0.35 |
ENST00000508423.1
|
KIAA0232
|
KIAA0232 |
| chr21_-_35014027 | 0.35 |
ENST00000399442.1
ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr1_+_40627038 | 0.35 |
ENST00000372771.4
|
RLF
|
rearranged L-myc fusion |
| chr13_-_31736132 | 0.35 |
ENST00000429785.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr2_+_170550944 | 0.35 |
ENST00000359744.3
ENST00000438838.1 ENST00000438710.1 ENST00000449906.1 ENST00000498202.2 ENST00000272797.4 |
PHOSPHO2
KLHL23
|
phosphatase, orphan 2 kelch-like family member 23 |
| chr6_+_142468383 | 0.34 |
ENST00000367621.1
ENST00000452973.2 |
VTA1
|
vesicle (multivesicular body) trafficking 1 |
| chr4_-_2420335 | 0.34 |
ENST00000503000.1
|
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr15_-_101142362 | 0.34 |
ENST00000559577.1
ENST00000561308.1 ENST00000560133.1 ENST00000560941.1 ENST00000559736.1 ENST00000560272.1 |
LINS
|
lines homolog (Drosophila) |
| chr1_+_206809113 | 0.34 |
ENST00000441486.1
ENST00000367106.1 |
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chrX_+_133371077 | 0.32 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr8_+_94767109 | 0.31 |
ENST00000409623.3
ENST00000453906.1 ENST00000521517.1 |
TMEM67
|
transmembrane protein 67 |
| chr7_-_129845313 | 0.31 |
ENST00000397622.2
|
TMEM209
|
transmembrane protein 209 |
| chr11_+_113185571 | 0.31 |
ENST00000455306.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr7_-_129845188 | 0.31 |
ENST00000462753.1
ENST00000471077.1 ENST00000473456.1 ENST00000336804.8 |
TMEM209
|
transmembrane protein 209 |
| chr3_+_52740094 | 0.31 |
ENST00000602728.1
|
SPCS1
|
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
| chr14_+_96343100 | 0.31 |
ENST00000503525.2
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr8_+_10530155 | 0.30 |
ENST00000521818.1
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr2_+_26785409 | 0.30 |
ENST00000329615.3
ENST00000409392.1 |
C2orf70
|
chromosome 2 open reading frame 70 |
| chr1_+_74663896 | 0.30 |
ENST00000370898.3
ENST00000467578.2 ENST00000370894.5 ENST00000482102.2 ENST00000609362.1 ENST00000534056.1 ENST00000557284.2 ENST00000370899.3 ENST00000370895.1 ENST00000534632.1 ENST00000370893.1 ENST00000370891.2 |
FPGT
FPGT-TNNI3K
TNNI3K
|
fucose-1-phosphate guanylyltransferase FPGT-TNNI3K readthrough TNNI3 interacting kinase |
| chr21_-_43916296 | 0.30 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr11_+_124543694 | 0.30 |
ENST00000227135.2
ENST00000532692.1 |
SPA17
|
sperm autoantigenic protein 17 |
| chr16_+_57406437 | 0.30 |
ENST00000564948.1
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1 |
| chr4_+_123653882 | 0.30 |
ENST00000433287.1
|
BBS12
|
Bardet-Biedl syndrome 12 |
| chr1_+_206808868 | 0.29 |
ENST00000367109.2
|
DYRK3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr1_-_38061522 | 0.29 |
ENST00000373062.3
|
GNL2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr14_-_22005062 | 0.29 |
ENST00000317492.5
|
SALL2
|
spalt-like transcription factor 2 |
| chr15_+_49913175 | 0.29 |
ENST00000403028.3
|
DTWD1
|
DTW domain containing 1 |
| chr1_+_39456895 | 0.29 |
ENST00000432648.3
ENST00000446189.2 ENST00000372984.4 |
AKIRIN1
|
akirin 1 |
| chr6_+_17281802 | 0.29 |
ENST00000509686.1
|
RBM24
|
RNA binding motif protein 24 |
| chr15_+_74610894 | 0.28 |
ENST00000558821.1
ENST00000268082.4 |
CCDC33
|
coiled-coil domain containing 33 |
| chr1_+_45140360 | 0.28 |
ENST00000418644.1
ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228
|
chromosome 1 open reading frame 228 |
| chr14_-_64846033 | 0.28 |
ENST00000556556.1
|
CTD-2555O16.1
|
CTD-2555O16.1 |
| chr15_-_55700457 | 0.28 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr4_-_156297919 | 0.28 |
ENST00000450097.1
|
MAP9
|
microtubule-associated protein 9 |
| chr15_-_101142401 | 0.28 |
ENST00000314742.8
|
LINS
|
lines homolog (Drosophila) |
| chr11_-_113577014 | 0.28 |
ENST00000544634.1
ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr3_-_130465604 | 0.27 |
ENST00000356763.3
|
PIK3R4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr2_-_47143160 | 0.27 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr8_-_93978333 | 0.27 |
ENST00000524037.1
ENST00000520430.1 ENST00000521617.1 |
TRIQK
|
triple QxxK/R motif containing |
| chr17_-_64187973 | 0.27 |
ENST00000583358.1
ENST00000392769.2 |
CEP112
|
centrosomal protein 112kDa |
| chr1_-_86861936 | 0.27 |
ENST00000394733.2
ENST00000359242.3 ENST00000294678.2 ENST00000479890.1 ENST00000317336.7 ENST00000370567.1 ENST00000394731.1 ENST00000478286.2 ENST00000370566.3 |
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr12_-_108154705 | 0.27 |
ENST00000547188.1
|
PRDM4
|
PR domain containing 4 |
| chr11_-_113577052 | 0.27 |
ENST00000540540.1
ENST00000545579.1 ENST00000538955.1 ENST00000299882.5 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr1_+_43613612 | 0.27 |
ENST00000335282.4
|
FAM183A
|
family with sequence similarity 183, member A |
| chr6_-_46620522 | 0.26 |
ENST00000275016.2
|
CYP39A1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr9_+_26956474 | 0.26 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr19_+_10217364 | 0.26 |
ENST00000430370.1
|
PPAN
|
peter pan homolog (Drosophila) |
| chr6_+_46620676 | 0.26 |
ENST00000371347.5
ENST00000411689.2 |
SLC25A27
|
solute carrier family 25, member 27 |
| chr3_-_195076933 | 0.26 |
ENST00000423531.1
|
ACAP2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr2_-_37458749 | 0.26 |
ENST00000234170.5
|
CEBPZ
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr11_-_8954491 | 0.26 |
ENST00000526227.1
ENST00000525780.1 ENST00000326053.5 |
C11orf16
|
chromosome 11 open reading frame 16 |
| chr20_-_49575058 | 0.25 |
ENST00000371584.4
ENST00000371583.5 ENST00000413082.1 |
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr12_+_93861282 | 0.25 |
ENST00000552217.1
ENST00000393128.4 ENST00000547098.1 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr3_+_158288942 | 0.25 |
ENST00000491767.1
ENST00000355893.5 |
MLF1
|
myeloid leukemia factor 1 |
| chr11_+_36616355 | 0.25 |
ENST00000532470.2
|
C11orf74
|
chromosome 11 open reading frame 74 |
| chr17_+_39846114 | 0.25 |
ENST00000586699.1
|
EIF1
|
eukaryotic translation initiation factor 1 |
| chr21_+_34602680 | 0.25 |
ENST00000447980.1
|
IFNAR2
|
interferon (alpha, beta and omega) receptor 2 |
| chr13_-_44735393 | 0.25 |
ENST00000400419.1
|
SMIM2
|
small integral membrane protein 2 |
| chr19_-_13710678 | 0.25 |
ENST00000592864.1
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr1_+_87380299 | 0.24 |
ENST00000370551.4
ENST00000370550.5 |
HS2ST1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr15_-_55700522 | 0.24 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr10_+_82116529 | 0.24 |
ENST00000411538.1
ENST00000256039.2 |
DYDC2
|
DPY30 domain containing 2 |
| chr19_-_3801789 | 0.24 |
ENST00000590849.1
ENST00000395045.2 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr2_-_153032484 | 0.24 |
ENST00000263904.4
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr5_-_76916396 | 0.24 |
ENST00000509971.1
|
WDR41
|
WD repeat domain 41 |
| chr2_+_196521845 | 0.24 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr1_+_43613566 | 0.24 |
ENST00000409396.1
|
FAM183A
|
family with sequence similarity 183, member A |
| chr15_-_66084621 | 0.24 |
ENST00000564674.1
|
DENND4A
|
DENN/MADD domain containing 4A |
| chr6_-_99963286 | 0.24 |
ENST00000369233.2
ENST00000500704.2 ENST00000329966.6 |
USP45
|
ubiquitin specific peptidase 45 |
| chr8_+_10530133 | 0.23 |
ENST00000304519.5
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chr2_-_62081254 | 0.23 |
ENST00000405894.3
|
FAM161A
|
family with sequence similarity 161, member A |
| chr15_+_49447947 | 0.23 |
ENST00000327171.3
ENST00000560654.1 |
GALK2
|
galactokinase 2 |
| chr6_-_99963252 | 0.23 |
ENST00000392738.2
ENST00000327681.6 ENST00000472914.2 |
USP45
|
ubiquitin specific peptidase 45 |
| chr13_-_31736478 | 0.23 |
ENST00000445273.2
|
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr18_-_54318353 | 0.23 |
ENST00000590954.1
ENST00000540155.1 |
TXNL1
|
thioredoxin-like 1 |
| chr10_-_97453650 | 0.23 |
ENST00000371209.5
ENST00000371217.5 ENST00000430368.2 |
TCTN3
|
tectonic family member 3 |
| chr7_-_36406750 | 0.23 |
ENST00000453212.1
ENST00000415803.2 ENST00000440378.1 ENST00000431396.1 ENST00000317020.6 ENST00000436884.1 |
KIAA0895
|
KIAA0895 |
| chr13_-_31736027 | 0.23 |
ENST00000380406.5
ENST00000320027.5 ENST00000380405.4 |
HSPH1
|
heat shock 105kDa/110kDa protein 1 |
| chr11_-_118122996 | 0.22 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr1_+_174969262 | 0.22 |
ENST00000406752.1
ENST00000405362.1 |
CACYBP
|
calcyclin binding protein |
| chr7_+_23637118 | 0.22 |
ENST00000448353.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr10_+_23216944 | 0.22 |
ENST00000298032.5
ENST00000409983.3 ENST00000409049.3 |
ARMC3
|
armadillo repeat containing 3 |
| chr5_-_110074603 | 0.22 |
ENST00000515278.2
|
TMEM232
|
transmembrane protein 232 |
| chr16_-_3545424 | 0.22 |
ENST00000437192.3
ENST00000399645.3 |
C16orf90
|
chromosome 16 open reading frame 90 |
| chr12_+_95611569 | 0.22 |
ENST00000261219.6
ENST00000551472.1 ENST00000552821.1 |
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr1_+_93646238 | 0.22 |
ENST00000448243.1
ENST00000370276.1 |
CCDC18
|
coiled-coil domain containing 18 |
| chr14_+_75536280 | 0.22 |
ENST00000238686.8
|
ZC2HC1C
|
zinc finger, C2HC-type containing 1C |
| chr11_+_113185292 | 0.22 |
ENST00000429951.1
ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12
|
tetratricopeptide repeat domain 12 |
| chr8_-_101118185 | 0.22 |
ENST00000523437.1
|
RGS22
|
regulator of G-protein signaling 22 |
| chr16_+_20912382 | 0.22 |
ENST00000396052.2
|
LYRM1
|
LYR motif containing 1 |
| chr16_+_20912075 | 0.21 |
ENST00000219168.4
|
LYRM1
|
LYR motif containing 1 |
| chr4_+_141445311 | 0.21 |
ENST00000323570.3
ENST00000511887.2 |
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr12_+_100661156 | 0.21 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr17_-_64188177 | 0.21 |
ENST00000535342.2
|
CEP112
|
centrosomal protein 112kDa |
| chr9_+_130478345 | 0.21 |
ENST00000373289.3
ENST00000393748.4 |
TTC16
|
tetratricopeptide repeat domain 16 |
| chr2_+_27851863 | 0.21 |
ENST00000264718.3
ENST00000610189.1 |
GPN1
|
GPN-loop GTPase 1 |
| chr5_-_137475071 | 0.21 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr16_-_71598823 | 0.20 |
ENST00000566202.1
|
ZNF19
|
zinc finger protein 19 |
| chr1_-_85155939 | 0.20 |
ENST00000603677.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr13_-_96705624 | 0.20 |
ENST00000376747.3
ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr3_-_121468602 | 0.20 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr1_-_119682812 | 0.20 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chrX_+_47444613 | 0.20 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr6_-_159065741 | 0.20 |
ENST00000367085.3
ENST00000367089.3 |
DYNLT1
|
dynein, light chain, Tctex-type 1 |
| chr14_+_96342729 | 0.20 |
ENST00000504119.1
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr16_+_50059182 | 0.20 |
ENST00000562576.1
|
CNEP1R1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr1_-_169396646 | 0.20 |
ENST00000367806.3
|
CCDC181
|
coiled-coil domain containing 181 |
| chr11_+_36616044 | 0.20 |
ENST00000334307.5
ENST00000531554.1 ENST00000347206.4 ENST00000534635.1 ENST00000446510.2 ENST00000530697.1 ENST00000527108.1 |
C11orf74
|
chromosome 11 open reading frame 74 |
| chr15_+_90728145 | 0.20 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr9_-_135545380 | 0.20 |
ENST00000544003.1
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr1_+_114473350 | 0.19 |
ENST00000503968.1
|
HIPK1
|
homeodomain interacting protein kinase 1 |
| chr2_+_183580954 | 0.19 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr6_+_89790459 | 0.19 |
ENST00000369472.1
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr3_+_158288999 | 0.19 |
ENST00000482628.1
ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1
|
myeloid leukemia factor 1 |
| chr5_+_121297650 | 0.19 |
ENST00000339397.4
|
SRFBP1
|
serum response factor binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX7_RFX4_RFX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 1.0 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 0.8 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.2 | 2.3 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.2 | 1.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 3.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.4 | GO:0051040 | regulation of calcium-independent cell-cell adhesion(GO:0051040) |
| 0.1 | 1.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.3 | GO:0039008 | pronephric nephron morphogenesis(GO:0039007) pronephric nephron tubule morphogenesis(GO:0039008) pronephric duct development(GO:0039022) pronephric duct morphogenesis(GO:0039023) Kupffer's vesicle development(GO:0070121) |
| 0.1 | 0.4 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.3 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.1 | 0.3 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.1 | 0.7 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.8 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.4 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.5 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 0.7 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.6 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.8 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.2 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.8 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.2 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.1 | 0.3 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 0.6 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 0.2 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.4 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.2 | GO:0061570 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.1 | 1.2 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) |
| 0.1 | 0.2 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.2 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 0.8 | GO:1904903 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.1 | 0.5 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 1.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 2.6 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.4 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 1.0 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.3 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 5.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.3 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0010983 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.4 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.3 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.1 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.0 | GO:0060298 | sarcomerogenesis(GO:0048769) positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.1 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 0.4 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0035811 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.0 | GO:0061528 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.5 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.2 | 0.7 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.4 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.1 | 1.8 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 3.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.2 | GO:0005668 | RNA polymerase transcription factor SL1 complex(GO:0005668) |
| 0.1 | 1.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) glial limiting end-foot(GO:0097451) |
| 0.0 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.3 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0036126 | sperm flagellum(GO:0036126) sperm part(GO:0097223) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.7 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 5.0 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.0 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.0 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.2 | 0.6 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.2 | 0.7 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 1.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.5 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.5 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.1 | 0.6 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.4 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.1 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.7 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.2 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.2 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.7 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 1.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) ATP-dependent helicase activity(GO:0008026) RNA-dependent ATPase activity(GO:0008186) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.2 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0023023 | MHC protein complex binding(GO:0023023) MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.0 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.4 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.0 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.2 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |