Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
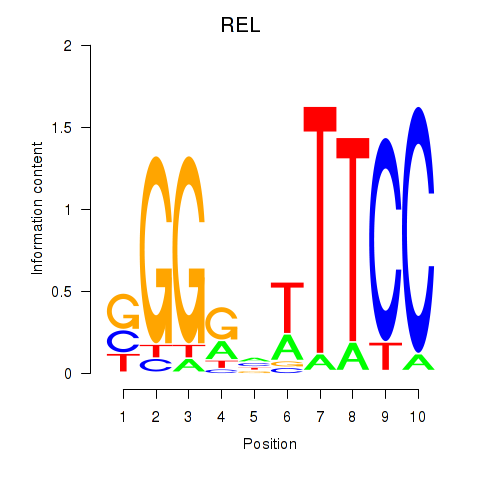
Results for REL
Z-value: 0.83
Transcription factors associated with REL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REL
|
ENSG00000162924.9 | REL proto-oncogene, NF-kB subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| REL | hg19_v2_chr2_+_61108771_61108791 | 0.46 | 3.6e-01 | Click! |
Activity profile of REL motif
Sorted Z-values of REL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_46546406 | 1.18 |
ENST00000440343.1
ENST00000415785.1 |
PPARA
|
peroxisome proliferator-activated receptor alpha |
| chr1_+_6845578 | 0.75 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr19_+_58193388 | 0.61 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
AC003006.7
|
zinc finger protein 551 Uncharacterized protein |
| chr15_+_44719996 | 0.54 |
ENST00000559793.1
ENST00000558968.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr17_-_15469590 | 0.53 |
ENST00000312127.2
|
CDRT1
|
CMT duplicated region transcript 1; Uncharacterized protein |
| chr20_-_36156264 | 0.52 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr22_-_28490123 | 0.45 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr19_-_4338783 | 0.45 |
ENST00000601482.1
ENST00000600324.1 |
STAP2
|
signal transducing adaptor family member 2 |
| chr6_-_30080863 | 0.43 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr4_-_185395191 | 0.42 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr3_+_169629354 | 0.41 |
ENST00000428432.2
ENST00000335556.3 |
SAMD7
|
sterile alpha motif domain containing 7 |
| chr6_+_138188351 | 0.41 |
ENST00000421450.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr16_+_84209539 | 0.41 |
ENST00000569735.1
|
DNAAF1
|
dynein, axonemal, assembly factor 1 |
| chr14_-_103589246 | 0.41 |
ENST00000558224.1
ENST00000560742.1 |
LINC00677
|
long intergenic non-protein coding RNA 677 |
| chr4_+_74702214 | 0.41 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chr2_-_191885686 | 0.40 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr1_+_6845497 | 0.40 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr6_-_30080876 | 0.38 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr2_+_208394658 | 0.36 |
ENST00000421139.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr22_+_22676808 | 0.36 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr4_+_74735102 | 0.32 |
ENST00000395761.3
|
CXCL1
|
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr14_+_37667230 | 0.32 |
ENST00000556451.1
ENST00000556753.1 ENST00000396294.2 |
MIPOL1
|
mirror-image polydactyly 1 |
| chr17_-_4642429 | 0.31 |
ENST00000573123.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr12_-_95009837 | 0.31 |
ENST00000551457.1
|
TMCC3
|
transmembrane and coiled-coil domain family 3 |
| chr2_+_208394616 | 0.31 |
ENST00000432329.2
ENST00000353267.3 ENST00000445803.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr1_-_205912577 | 0.30 |
ENST00000367135.3
ENST00000367134.2 |
SLC26A9
|
solute carrier family 26 (anion exchanger), member 9 |
| chr4_+_185395947 | 0.30 |
ENST00000605834.1
|
RP11-326I11.3
|
RP11-326I11.3 |
| chr5_+_118604439 | 0.27 |
ENST00000388882.5
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr9_+_101705893 | 0.27 |
ENST00000375001.3
|
COL15A1
|
collagen, type XV, alpha 1 |
| chr10_+_89124746 | 0.27 |
ENST00000465545.1
|
NUTM2D
|
NUT family member 2D |
| chr13_-_88323514 | 0.26 |
ENST00000441617.1
|
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr10_+_30723105 | 0.26 |
ENST00000375322.2
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr8_-_101321584 | 0.26 |
ENST00000523167.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr5_-_127418573 | 0.25 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr19_+_58193337 | 0.25 |
ENST00000601064.1
ENST00000282296.5 ENST00000356715.4 |
ZNF551
|
zinc finger protein 551 |
| chr2_-_113594279 | 0.25 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr3_-_179322436 | 0.25 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr2_-_204400113 | 0.25 |
ENST00000319170.5
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr10_-_91295304 | 0.25 |
ENST00000341233.4
ENST00000371790.4 |
SLC16A12
|
solute carrier family 16, member 12 |
| chr11_+_46316677 | 0.25 |
ENST00000534787.1
|
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr20_+_43803517 | 0.22 |
ENST00000243924.3
|
PI3
|
peptidase inhibitor 3, skin-derived |
| chr7_-_99679324 | 0.22 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr7_-_102252589 | 0.22 |
ENST00000520042.1
|
RASA4
|
RAS p21 protein activator 4 |
| chr11_+_102188272 | 0.20 |
ENST00000532808.1
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chr2_-_128145498 | 0.20 |
ENST00000409179.2
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr7_+_107384579 | 0.20 |
ENST00000222597.2
ENST00000415884.2 |
CBLL1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr6_-_143832820 | 0.20 |
ENST00000002165.6
|
FUCA2
|
fucosidase, alpha-L- 2, plasma |
| chr4_-_176733897 | 0.20 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr10_+_90750493 | 0.19 |
ENST00000357339.2
ENST00000355279.2 |
FAS
|
Fas cell surface death receptor |
| chr19_-_49250054 | 0.19 |
ENST00000602105.1
ENST00000332955.2 |
IZUMO1
|
izumo sperm-egg fusion 1 |
| chr2_+_208394794 | 0.19 |
ENST00000536726.1
ENST00000374397.4 ENST00000452474.1 |
CREB1
|
cAMP responsive element binding protein 1 |
| chr15_+_33010175 | 0.19 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr11_+_102188224 | 0.19 |
ENST00000263464.3
|
BIRC3
|
baculoviral IAP repeat containing 3 |
| chrX_-_20284733 | 0.19 |
ENST00000438357.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr1_-_62190793 | 0.19 |
ENST00000371177.2
ENST00000606498.1 |
TM2D1
|
TM2 domain containing 1 |
| chr2_+_238600933 | 0.19 |
ENST00000420665.1
ENST00000392000.4 |
LRRFIP1
|
leucine rich repeat (in FLII) interacting protein 1 |
| chr12_-_86230315 | 0.19 |
ENST00000361228.3
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
| chr3_-_179322416 | 0.18 |
ENST00000259038.2
|
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr4_-_74904398 | 0.17 |
ENST00000296026.4
|
CXCL3
|
chemokine (C-X-C motif) ligand 3 |
| chr4_+_114214125 | 0.17 |
ENST00000509550.1
|
ANK2
|
ankyrin 2, neuronal |
| chr2_+_202047843 | 0.17 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr11_-_75062730 | 0.16 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr4_-_146101304 | 0.16 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chr3_+_52719936 | 0.16 |
ENST00000418458.1
ENST00000394799.2 |
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr5_-_59064458 | 0.16 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr10_+_30723045 | 0.15 |
ENST00000542547.1
ENST00000415139.1 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr19_-_39390212 | 0.15 |
ENST00000437828.1
|
SIRT2
|
sirtuin 2 |
| chr11_-_75062829 | 0.15 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr4_-_76957214 | 0.15 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr3_+_197518100 | 0.15 |
ENST00000438796.2
ENST00000414675.2 ENST00000441090.2 ENST00000334859.4 ENST00000425562.2 |
LRCH3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr10_-_6104253 | 0.15 |
ENST00000256876.6
ENST00000379954.1 ENST00000379959.3 |
IL2RA
|
interleukin 2 receptor, alpha |
| chr2_-_61108449 | 0.15 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr2_-_113993020 | 0.15 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr1_-_23694794 | 0.15 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr13_+_95364963 | 0.14 |
ENST00000438290.2
|
SOX21-AS1
|
SOX21 antisense RNA 1 (head to head) |
| chr18_+_3252265 | 0.14 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_+_102271129 | 0.14 |
ENST00000258534.8
|
DRAM1
|
DNA-damage regulated autophagy modulator 1 |
| chr22_+_46731676 | 0.14 |
ENST00000424260.2
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr12_-_56211485 | 0.14 |
ENST00000552080.1
ENST00000444631.2 ENST00000336133.3 |
SARNP
|
SAP domain containing ribonucleoprotein |
| chr8_+_144816303 | 0.14 |
ENST00000533004.1
|
FAM83H-AS1
|
FAM83H antisense RNA 1 (head to head) |
| chr1_+_95582881 | 0.14 |
ENST00000370203.4
ENST00000456991.1 |
TMEM56
|
transmembrane protein 56 |
| chr15_+_71145578 | 0.14 |
ENST00000544974.2
ENST00000558546.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr22_+_46731596 | 0.13 |
ENST00000381019.3
|
TRMU
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr10_+_22614547 | 0.13 |
ENST00000416820.1
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr1_-_27216729 | 0.13 |
ENST00000431781.2
ENST00000374135.4 |
GPN2
|
GPN-loop GTPase 2 |
| chr3_+_184081137 | 0.13 |
ENST00000443489.1
|
POLR2H
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr2_+_204193101 | 0.13 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chr10_-_46970533 | 0.13 |
ENST00000512997.1
|
SYT15
|
synaptotagmin XV |
| chr2_+_202047596 | 0.13 |
ENST00000286186.6
ENST00000360132.3 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr4_-_74864386 | 0.13 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr2_-_43453734 | 0.13 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr2_+_208394455 | 0.13 |
ENST00000430624.1
|
CREB1
|
cAMP responsive element binding protein 1 |
| chr6_+_30850862 | 0.12 |
ENST00000504651.1
ENST00000512694.1 ENST00000515233.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr17_-_8286484 | 0.12 |
ENST00000582556.1
ENST00000584164.1 ENST00000293842.5 ENST00000584343.1 ENST00000578812.1 ENST00000583011.1 |
RPL26
|
ribosomal protein L26 |
| chr3_-_139108475 | 0.12 |
ENST00000515006.1
ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr20_-_44937124 | 0.12 |
ENST00000537909.1
|
CDH22
|
cadherin 22, type 2 |
| chr19_+_11466062 | 0.12 |
ENST00000251473.5
ENST00000591329.1 ENST00000586380.1 |
DKFZP761J1410
|
Lipid phosphate phosphatase-related protein type 2 |
| chr6_-_166075557 | 0.12 |
ENST00000539869.2
ENST00000366882.1 |
PDE10A
|
phosphodiesterase 10A |
| chr2_-_88355241 | 0.12 |
ENST00000347055.3
|
KRCC1
|
lysine-rich coiled-coil 1 |
| chr3_+_141596371 | 0.12 |
ENST00000495216.1
|
ATP1B3
|
ATPase, Na+/K+ transporting, beta 3 polypeptide |
| chr8_+_42128861 | 0.12 |
ENST00000518983.1
|
IKBKB
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr6_-_32122106 | 0.11 |
ENST00000428778.1
|
PRRT1
|
proline-rich transmembrane protein 1 |
| chr22_-_41252962 | 0.11 |
ENST00000216218.3
|
ST13
|
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
| chr11_-_64570706 | 0.11 |
ENST00000294066.2
ENST00000377350.3 |
MAP4K2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr12_-_53297432 | 0.11 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr14_+_77648167 | 0.11 |
ENST00000554346.1
ENST00000298351.4 |
TMEM63C
|
transmembrane protein 63C |
| chr17_-_73178599 | 0.11 |
ENST00000578238.1
|
SUMO2
|
small ubiquitin-like modifier 2 |
| chr2_+_228189941 | 0.11 |
ENST00000353339.3
ENST00000354503.6 ENST00000530359.1 ENST00000531278.1 ENST00000409565.1 ENST00000452930.1 ENST00000409616.1 ENST00000337110.7 ENST00000525195.1 ENST00000534203.1 ENST00000524634.1 ENST00000349901.7 |
MFF
|
mitochondrial fission factor |
| chr10_-_105437909 | 0.11 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr14_+_68086515 | 0.11 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr12_-_96793142 | 0.11 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr14_-_35873856 | 0.11 |
ENST00000553342.1
ENST00000216797.5 ENST00000557140.1 |
NFKBIA
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr10_+_30722866 | 0.11 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr2_+_102314161 | 0.11 |
ENST00000425019.1
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr9_-_115480303 | 0.11 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chr17_-_45266542 | 0.10 |
ENST00000531206.1
ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27
|
cell division cycle 27 |
| chr12_-_77272765 | 0.10 |
ENST00000547435.1
ENST00000552330.1 ENST00000546966.1 ENST00000311083.5 |
CSRP2
|
cysteine and glycine-rich protein 2 |
| chr2_-_18741882 | 0.10 |
ENST00000381249.3
|
RDH14
|
retinol dehydrogenase 14 (all-trans/9-cis/11-cis) |
| chr21_-_34915147 | 0.10 |
ENST00000381831.3
ENST00000381839.3 |
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr1_-_35658736 | 0.10 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr18_+_33877654 | 0.10 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr11_-_72852958 | 0.10 |
ENST00000458644.2
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr9_-_13279406 | 0.10 |
ENST00000546205.1
|
MPDZ
|
multiple PDZ domain protein |
| chr3_-_119379719 | 0.10 |
ENST00000493094.1
|
POPDC2
|
popeye domain containing 2 |
| chr19_-_17958771 | 0.10 |
ENST00000534444.1
|
JAK3
|
Janus kinase 3 |
| chr1_+_56880606 | 0.10 |
ENST00000451914.1
|
RP4-710M16.2
|
RP4-710M16.2 |
| chr11_+_46299199 | 0.10 |
ENST00000529193.1
ENST00000288400.3 |
CREB3L1
|
cAMP responsive element binding protein 3-like 1 |
| chr17_-_75878647 | 0.10 |
ENST00000374983.2
|
FLJ45079
|
FLJ45079 |
| chr19_+_35940486 | 0.10 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr14_-_24616426 | 0.09 |
ENST00000216802.5
|
PSME2
|
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr11_+_120255997 | 0.09 |
ENST00000532993.1
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr7_-_102715263 | 0.09 |
ENST00000379305.3
|
FBXL13
|
F-box and leucine-rich repeat protein 13 |
| chr1_-_8000872 | 0.09 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr9_+_127054254 | 0.09 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr13_-_52027134 | 0.09 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr9_-_34637806 | 0.09 |
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr17_+_40985407 | 0.09 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr1_-_226595648 | 0.09 |
ENST00000366790.3
|
PARP1
|
poly (ADP-ribose) polymerase 1 |
| chr19_-_6591113 | 0.09 |
ENST00000423145.3
ENST00000245903.3 |
CD70
|
CD70 molecule |
| chr16_+_50730910 | 0.09 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr16_+_3014217 | 0.09 |
ENST00000572045.1
|
KREMEN2
|
kringle containing transmembrane protein 2 |
| chr1_-_204380919 | 0.09 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr17_-_16118835 | 0.09 |
ENST00000582357.1
ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr6_+_30850697 | 0.09 |
ENST00000509639.1
ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_-_27816556 | 0.09 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr9_-_115095229 | 0.09 |
ENST00000210227.4
|
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr8_+_72755796 | 0.08 |
ENST00000524152.1
|
RP11-383H13.1
|
Protein LOC100132891; cDNA FLJ53548 |
| chr20_+_42984330 | 0.08 |
ENST00000316673.4
ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A
|
hepatocyte nuclear factor 4, alpha |
| chr2_+_220094657 | 0.08 |
ENST00000436226.1
|
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr21_-_34915084 | 0.08 |
ENST00000426819.1
|
GART
|
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr5_+_82767284 | 0.08 |
ENST00000265077.3
|
VCAN
|
versican |
| chr17_+_49337881 | 0.08 |
ENST00000225298.7
|
UTP18
|
UTP18 small subunit (SSU) processome component homolog (yeast) |
| chr12_-_57023995 | 0.08 |
ENST00000549884.1
ENST00000546695.1 |
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr5_-_141703713 | 0.08 |
ENST00000511815.1
|
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr18_+_21529811 | 0.08 |
ENST00000588004.1
|
LAMA3
|
laminin, alpha 3 |
| chr12_+_51632508 | 0.08 |
ENST00000449723.3
|
DAZAP2
|
DAZ associated protein 2 |
| chr6_-_32821599 | 0.08 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr15_-_51914810 | 0.07 |
ENST00000543779.2
ENST00000449909.3 |
DMXL2
|
Dmx-like 2 |
| chr4_-_185395672 | 0.07 |
ENST00000393593.3
|
IRF2
|
interferon regulatory factor 2 |
| chr9_+_130911770 | 0.07 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr2_+_204193129 | 0.07 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr17_-_6338472 | 0.07 |
ENST00000570466.1
ENST00000576776.1 ENST00000576307.1 ENST00000381129.3 ENST00000250087.5 |
AIPL1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr2_+_204192942 | 0.07 |
ENST00000295851.5
ENST00000261017.5 |
ABI2
|
abl-interactor 2 |
| chr16_-_15736953 | 0.07 |
ENST00000548025.1
ENST00000551742.1 ENST00000602337.1 ENST00000344181.3 ENST00000396368.3 |
KIAA0430
|
KIAA0430 |
| chr3_-_139108463 | 0.07 |
ENST00000512242.1
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr2_+_173940668 | 0.07 |
ENST00000375213.3
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr11_+_7597639 | 0.07 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr9_-_115095883 | 0.07 |
ENST00000450374.1
ENST00000374255.2 ENST00000334318.6 ENST00000374257.1 |
PTBP3
|
polypyrimidine tract binding protein 3 |
| chr11_-_72492903 | 0.07 |
ENST00000537947.1
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr19_-_47734448 | 0.07 |
ENST00000439096.2
|
BBC3
|
BCL2 binding component 3 |
| chr6_-_30710265 | 0.07 |
ENST00000438162.1
ENST00000454845.1 |
FLOT1
|
flotillin 1 |
| chr21_+_34775772 | 0.07 |
ENST00000405436.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr2_+_204193149 | 0.07 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr5_-_55290773 | 0.07 |
ENST00000502326.3
ENST00000381298.2 |
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr3_-_156878540 | 0.07 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr14_-_30396802 | 0.07 |
ENST00000415220.2
|
PRKD1
|
protein kinase D1 |
| chr3_+_179322573 | 0.07 |
ENST00000493866.1
ENST00000472629.1 ENST00000482604.1 |
NDUFB5
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr2_+_162016827 | 0.07 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr1_+_235530675 | 0.07 |
ENST00000366601.3
ENST00000406207.1 ENST00000543662.1 |
TBCE
|
tubulin folding cofactor E |
| chr9_+_130911723 | 0.07 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr12_-_117537240 | 0.06 |
ENST00000392545.4
ENST00000541210.1 ENST00000335209.7 |
TESC
|
tescalcin |
| chr14_+_37667193 | 0.06 |
ENST00000539062.2
|
MIPOL1
|
mirror-image polydactyly 1 |
| chr1_+_165796753 | 0.06 |
ENST00000367879.4
|
UCK2
|
uridine-cytidine kinase 2 |
| chr19_-_4338838 | 0.06 |
ENST00000594605.1
|
STAP2
|
signal transducing adaptor family member 2 |
| chr15_+_85923856 | 0.06 |
ENST00000560302.1
ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13
|
A kinase (PRKA) anchor protein 13 |
| chr19_+_39390320 | 0.06 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr2_+_201987200 | 0.06 |
ENST00000425030.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_-_151431647 | 0.06 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr11_-_64856497 | 0.06 |
ENST00000524632.1
ENST00000530719.1 |
AP003068.6
|
transmembrane protein 262 |
| chr2_-_163175133 | 0.06 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr4_-_134070250 | 0.06 |
ENST00000505289.1
ENST00000509715.1 |
RP11-9G1.3
|
RP11-9G1.3 |
| chr11_-_65381643 | 0.06 |
ENST00000309100.3
ENST00000529839.1 ENST00000526293.1 |
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11 |
| chr2_+_162016916 | 0.06 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr2_+_220094479 | 0.06 |
ENST00000323348.5
ENST00000453432.1 ENST00000409849.1 ENST00000416565.1 ENST00000410034.3 ENST00000447157.1 |
ANKZF1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr1_-_32229934 | 0.06 |
ENST00000398542.1
|
BAI2
|
brain-specific angiogenesis inhibitor 2 |
| chr11_+_65479462 | 0.06 |
ENST00000377046.3
ENST00000352980.4 ENST00000341318.4 |
KAT5
|
K(lysine) acetyltransferase 5 |
| chr19_-_41256207 | 0.06 |
ENST00000598485.2
ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54
|
chromosome 19 open reading frame 54 |
| chr11_+_119039069 | 0.06 |
ENST00000422249.1
|
NLRX1
|
NLR family member X1 |
| chr3_+_101280677 | 0.06 |
ENST00000309922.6
ENST00000495642.1 |
TRMT10C
|
tRNA methyltransferase 10 homolog C (S. cerevisiae) |
| chr11_+_63997750 | 0.06 |
ENST00000321685.3
|
DNAJC4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of REL
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0072361 | regulation of glycolytic process by regulation of transcription from RNA polymerase II promoter(GO:0072361) |
| 0.1 | 1.0 | GO:1904550 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 0.4 | GO:0046725 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.1 | 0.4 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.1 | 0.2 | GO:0015881 | creatine transport(GO:0015881) creatine transmembrane transport(GO:1902598) |
| 0.1 | 0.3 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.3 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.4 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.3 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0061433 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.1 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.2 | GO:2000535 | regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.6 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:1902231 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0035509 | negative regulation of myosin-light-chain-phosphatase activity(GO:0035509) negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 1.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0007620 | copulation(GO:0007620) |
| 0.0 | 0.2 | GO:0010818 | T cell chemotaxis(GO:0010818) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.5 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.0 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.1 | 0.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 1.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.2 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 0.2 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.1 | GO:0004644 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.0 | 0.1 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.7 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.3 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.4 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.8 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |