Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
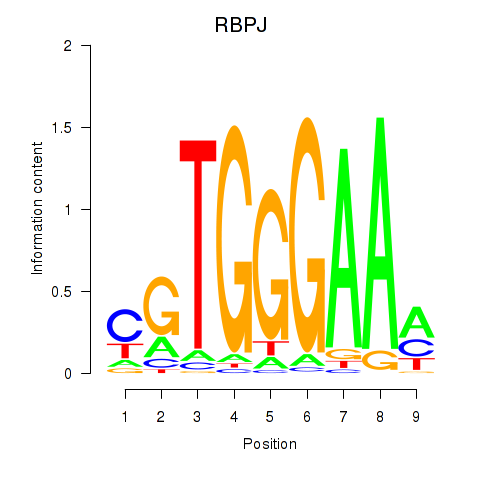
Results for RBPJ
Z-value: 0.49
Transcription factors associated with RBPJ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RBPJ
|
ENSG00000168214.16 | recombination signal binding protein for immunoglobulin kappa J region |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RBPJ | hg19_v2_chr4_+_26165074_26165110 | 0.57 | 2.4e-01 | Click! |
Activity profile of RBPJ motif
Sorted Z-values of RBPJ motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_38146965 | 0.28 |
ENST00000395873.3
ENST00000357328.4 ENST00000395874.2 |
ZNF248
|
zinc finger protein 248 |
| chr1_-_197744763 | 0.28 |
ENST00000422998.1
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr16_+_4666475 | 0.25 |
ENST00000591895.1
|
MGRN1
|
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr2_+_38177575 | 0.23 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr15_+_57891609 | 0.22 |
ENST00000569089.1
|
MYZAP
|
myocardial zonula adherens protein |
| chr11_-_790060 | 0.19 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr2_-_164592497 | 0.18 |
ENST00000333129.3
ENST00000409634.1 |
FIGN
|
fidgetin |
| chr8_-_80680078 | 0.18 |
ENST00000337919.5
ENST00000354724.3 |
HEY1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr3_+_171561127 | 0.18 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chrX_-_153095813 | 0.17 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chr1_-_243349684 | 0.17 |
ENST00000522895.1
|
CEP170
|
centrosomal protein 170kDa |
| chr10_-_38146482 | 0.16 |
ENST00000374648.3
|
ZNF248
|
zinc finger protein 248 |
| chr12_-_31945172 | 0.16 |
ENST00000340398.3
|
H3F3C
|
H3 histone, family 3C |
| chr1_-_197744390 | 0.13 |
ENST00000367396.3
|
DENND1B
|
DENN/MADD domain containing 1B |
| chr8_-_101348408 | 0.13 |
ENST00000519527.1
ENST00000522369.1 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr14_+_71788096 | 0.13 |
ENST00000557151.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr8_+_55466915 | 0.12 |
ENST00000522711.2
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr11_-_8290263 | 0.12 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr6_-_31612808 | 0.12 |
ENST00000438149.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr1_+_6511651 | 0.11 |
ENST00000434576.1
|
ESPN
|
espin |
| chr20_+_32250079 | 0.11 |
ENST00000375222.3
|
C20orf144
|
chromosome 20 open reading frame 144 |
| chr3_-_45957088 | 0.11 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr1_-_8000872 | 0.11 |
ENST00000377507.3
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr10_-_38146510 | 0.11 |
ENST00000395867.3
|
ZNF248
|
zinc finger protein 248 |
| chr1_-_935361 | 0.11 |
ENST00000484667.2
|
HES4
|
hes family bHLH transcription factor 4 |
| chrX_+_122993544 | 0.10 |
ENST00000422098.1
|
XIAP
|
X-linked inhibitor of apoptosis |
| chr15_-_70390213 | 0.10 |
ENST00000557997.1
ENST00000317509.8 ENST00000442299.2 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chrX_+_16141667 | 0.10 |
ENST00000380289.2
|
GRPR
|
gastrin-releasing peptide receptor |
| chr1_-_146644036 | 0.10 |
ENST00000425272.2
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr3_+_69812877 | 0.10 |
ENST00000457080.1
ENST00000328528.6 |
MITF
|
microphthalmia-associated transcription factor |
| chr5_+_63461642 | 0.10 |
ENST00000296615.6
ENST00000381081.2 ENST00000389100.4 |
RNF180
|
ring finger protein 180 |
| chr5_-_139943830 | 0.09 |
ENST00000412920.3
ENST00000511201.2 ENST00000356738.2 ENST00000354402.5 ENST00000358580.5 ENST00000508496.2 |
APBB3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr17_+_48351785 | 0.09 |
ENST00000507382.1
|
TMEM92
|
transmembrane protein 92 |
| chr3_+_32726774 | 0.09 |
ENST00000538368.1
|
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr19_+_19516561 | 0.08 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr17_-_62499334 | 0.08 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr1_-_85870177 | 0.08 |
ENST00000542148.1
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1 |
| chr6_+_144665237 | 0.08 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr13_-_25496926 | 0.08 |
ENST00000545981.1
ENST00000381884.4 |
CENPJ
|
centromere protein J |
| chr5_-_137071689 | 0.07 |
ENST00000505853.1
|
KLHL3
|
kelch-like family member 3 |
| chr2_+_208104351 | 0.07 |
ENST00000440326.1
|
AC007879.7
|
AC007879.7 |
| chr17_-_36891830 | 0.07 |
ENST00000578487.1
|
PCGF2
|
polycomb group ring finger 2 |
| chr11_+_7598239 | 0.07 |
ENST00000525597.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr6_+_126070726 | 0.07 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr19_+_7660716 | 0.07 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr2_+_187350973 | 0.07 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr19_+_44100632 | 0.07 |
ENST00000533118.1
|
ZNF576
|
zinc finger protein 576 |
| chr17_-_56606705 | 0.07 |
ENST00000317268.3
|
SEPT4
|
septin 4 |
| chr11_-_107436443 | 0.06 |
ENST00000429370.1
ENST00000417449.2 ENST00000428149.2 |
ALKBH8
|
alkB, alkylation repair homolog 8 (E. coli) |
| chr10_-_44144152 | 0.06 |
ENST00000395797.1
|
ZNF32
|
zinc finger protein 32 |
| chr2_+_187350883 | 0.06 |
ENST00000337859.6
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr3_+_46449049 | 0.06 |
ENST00000357392.4
ENST00000400880.3 ENST00000433848.1 |
CCRL2
|
chemokine (C-C motif) receptor-like 2 |
| chr11_+_64053311 | 0.06 |
ENST00000540370.1
|
GPR137
|
G protein-coupled receptor 137 |
| chr1_-_85358850 | 0.06 |
ENST00000370611.3
|
LPAR3
|
lysophosphatidic acid receptor 3 |
| chr4_-_176733897 | 0.06 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr11_-_101454658 | 0.06 |
ENST00000344327.3
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr7_+_6713376 | 0.06 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr18_+_66382428 | 0.06 |
ENST00000578970.1
ENST00000582371.1 ENST00000584775.1 |
CCDC102B
|
coiled-coil domain containing 102B |
| chr13_-_20357110 | 0.05 |
ENST00000427943.1
|
PSPC1
|
paraspeckle component 1 |
| chr2_-_129076151 | 0.05 |
ENST00000259241.6
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1 |
| chr17_-_70417365 | 0.05 |
ENST00000580948.1
|
LINC00511
|
long intergenic non-protein coding RNA 511 |
| chr9_+_88556036 | 0.05 |
ENST00000361671.5
ENST00000416045.1 |
NAA35
|
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr18_-_70305745 | 0.05 |
ENST00000581073.1
|
CBLN2
|
cerebellin 2 precursor |
| chr1_+_110026544 | 0.05 |
ENST00000369870.3
|
ATXN7L2
|
ataxin 7-like 2 |
| chr1_+_61547405 | 0.05 |
ENST00000371189.4
|
NFIA
|
nuclear factor I/A |
| chr8_+_55467072 | 0.05 |
ENST00000602362.1
|
RP11-53M11.3
|
RP11-53M11.3 |
| chr12_-_90049878 | 0.05 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chrX_-_49121165 | 0.05 |
ENST00000376207.4
ENST00000376199.2 |
FOXP3
|
forkhead box P3 |
| chr8_-_87242589 | 0.05 |
ENST00000419776.2
ENST00000297524.3 |
SLC7A13
|
solute carrier family 7 (anionic amino acid transporter), member 13 |
| chr1_+_955448 | 0.05 |
ENST00000379370.2
|
AGRN
|
agrin |
| chr17_+_29421987 | 0.05 |
ENST00000431387.4
|
NF1
|
neurofibromin 1 |
| chr12_-_14133053 | 0.05 |
ENST00000609686.1
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| chrX_-_118739835 | 0.05 |
ENST00000542113.1
ENST00000304449.5 |
NKRF
|
NFKB repressing factor |
| chr16_-_53737722 | 0.05 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr17_-_2239729 | 0.05 |
ENST00000576112.2
|
TSR1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr6_+_33176265 | 0.04 |
ENST00000374656.4
|
RING1
|
ring finger protein 1 |
| chr19_+_50529212 | 0.04 |
ENST00000270617.3
ENST00000445728.3 ENST00000601364.1 |
ZNF473
|
zinc finger protein 473 |
| chr11_+_45944190 | 0.04 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr7_+_102553430 | 0.04 |
ENST00000339431.4
ENST00000249377.4 |
LRRC17
|
leucine rich repeat containing 17 |
| chr11_+_66512089 | 0.04 |
ENST00000524551.1
ENST00000525908.1 ENST00000360962.4 ENST00000346672.4 ENST00000527634.1 ENST00000540737.1 |
C11orf80
|
chromosome 11 open reading frame 80 |
| chr8_+_21914477 | 0.04 |
ENST00000517804.1
|
DMTN
|
dematin actin binding protein |
| chr19_+_50528971 | 0.04 |
ENST00000598809.1
ENST00000595661.1 ENST00000391821.2 |
ZNF473
|
zinc finger protein 473 |
| chr1_+_82165350 | 0.04 |
ENST00000359929.3
|
LPHN2
|
latrophilin 2 |
| chr9_+_102584128 | 0.04 |
ENST00000338488.4
ENST00000395097.2 |
NR4A3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr19_-_17356697 | 0.04 |
ENST00000291442.3
|
NR2F6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr1_+_210406121 | 0.04 |
ENST00000367012.3
|
SERTAD4
|
SERTA domain containing 4 |
| chr16_-_70719925 | 0.04 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr12_-_90049828 | 0.04 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr10_-_44144292 | 0.04 |
ENST00000374433.2
|
ZNF32
|
zinc finger protein 32 |
| chr10_-_27530997 | 0.04 |
ENST00000375901.1
ENST00000412279.1 ENST00000375905.4 |
ACBD5
|
acyl-CoA binding domain containing 5 |
| chr7_+_128784712 | 0.04 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr18_+_10526008 | 0.04 |
ENST00000542979.1
ENST00000322897.6 |
NAPG
|
N-ethylmaleimide-sensitive factor attachment protein, gamma |
| chr1_+_15272271 | 0.04 |
ENST00000400797.3
|
KAZN
|
kazrin, periplakin interacting protein |
| chr19_-_40724246 | 0.04 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr6_-_31613280 | 0.04 |
ENST00000453833.1
|
BAG6
|
BCL2-associated athanogene 6 |
| chr13_+_39612442 | 0.03 |
ENST00000470258.1
ENST00000379600.3 |
NHLRC3
|
NHL repeat containing 3 |
| chr16_-_53737795 | 0.03 |
ENST00000262135.4
ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L
|
RPGRIP1-like |
| chr3_-_183735651 | 0.03 |
ENST00000427120.2
ENST00000392579.2 ENST00000382494.2 ENST00000446941.2 |
ABCC5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr16_+_85645007 | 0.03 |
ENST00000405402.2
|
GSE1
|
Gse1 coiled-coil protein |
| chr14_-_53258314 | 0.03 |
ENST00000216410.3
ENST00000557604.1 |
GNPNAT1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr14_+_51955831 | 0.03 |
ENST00000356218.4
|
FRMD6
|
FERM domain containing 6 |
| chr16_+_75182376 | 0.03 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chr17_-_56606664 | 0.03 |
ENST00000580844.1
|
SEPT4
|
septin 4 |
| chr19_+_56652556 | 0.03 |
ENST00000337080.3
|
ZNF444
|
zinc finger protein 444 |
| chr4_-_73935409 | 0.03 |
ENST00000507544.2
ENST00000295890.4 |
COX18
|
COX18 cytochrome C oxidase assembly factor |
| chr1_-_235667716 | 0.03 |
ENST00000313984.3
ENST00000366600.3 |
B3GALNT2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr7_+_76090993 | 0.03 |
ENST00000425780.1
ENST00000456590.1 ENST00000451769.1 ENST00000324432.5 ENST00000307569.8 ENST00000457529.1 ENST00000446600.1 ENST00000413936.2 ENST00000423646.1 ENST00000438930.1 ENST00000430490.2 |
DTX2
|
deltex homolog 2 (Drosophila) |
| chr17_-_56606639 | 0.03 |
ENST00000579371.1
|
SEPT4
|
septin 4 |
| chrX_+_106045891 | 0.03 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr10_-_51371321 | 0.03 |
ENST00000602930.1
|
AGAP8
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8 |
| chr19_+_39390320 | 0.03 |
ENST00000576510.1
|
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr14_+_24701628 | 0.03 |
ENST00000355299.4
ENST00000559836.1 |
GMPR2
|
guanosine monophosphate reductase 2 |
| chrX_-_153236620 | 0.03 |
ENST00000369984.4
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr2_-_145188137 | 0.03 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr5_-_115872142 | 0.03 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr2_+_208104497 | 0.03 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr1_+_113933371 | 0.03 |
ENST00000369617.4
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
| chr3_-_19975665 | 0.03 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr17_+_73452545 | 0.02 |
ENST00000314256.7
|
KIAA0195
|
KIAA0195 |
| chr11_+_123430948 | 0.02 |
ENST00000529432.1
ENST00000534764.1 |
GRAMD1B
|
GRAM domain containing 1B |
| chr13_-_39612176 | 0.02 |
ENST00000352251.3
ENST00000350125.3 |
PROSER1
|
proline and serine rich 1 |
| chr1_+_61869748 | 0.02 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr4_+_26321284 | 0.02 |
ENST00000506956.1
ENST00000512671.1 ENST00000345843.3 ENST00000342295.1 |
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr17_+_38474489 | 0.02 |
ENST00000394089.2
ENST00000425707.3 |
RARA
|
retinoic acid receptor, alpha |
| chr5_+_132009675 | 0.02 |
ENST00000231449.2
ENST00000350025.2 |
IL4
|
interleukin 4 |
| chr13_+_39612485 | 0.02 |
ENST00000379599.2
|
NHLRC3
|
NHL repeat containing 3 |
| chrX_-_153236819 | 0.02 |
ENST00000354233.3
|
HCFC1
|
host cell factor C1 (VP16-accessory protein) |
| chr19_+_56652686 | 0.02 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr19_+_44100727 | 0.02 |
ENST00000528387.1
ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576
SRRM5
|
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr10_-_33623826 | 0.02 |
ENST00000374867.2
|
NRP1
|
neuropilin 1 |
| chr7_-_120498357 | 0.02 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chrX_+_123094369 | 0.02 |
ENST00000455404.1
ENST00000218089.9 |
STAG2
|
stromal antigen 2 |
| chr3_+_142442841 | 0.02 |
ENST00000476941.1
ENST00000273482.6 |
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr1_-_211752073 | 0.02 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr3_-_100712352 | 0.02 |
ENST00000471714.1
ENST00000284322.5 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chrX_-_40594755 | 0.02 |
ENST00000324817.1
|
MED14
|
mediator complex subunit 14 |
| chr3_+_32726620 | 0.01 |
ENST00000331889.6
ENST00000328834.5 |
CNOT10
|
CCR4-NOT transcription complex, subunit 10 |
| chr10_-_128110441 | 0.01 |
ENST00000456514.1
|
LINC00601
|
long intergenic non-protein coding RNA 601 |
| chr1_+_161676739 | 0.01 |
ENST00000236938.6
ENST00000367959.2 ENST00000546024.1 ENST00000540521.1 ENST00000367949.2 ENST00000350710.3 ENST00000540926.1 |
FCRLA
|
Fc receptor-like A |
| chr20_+_30407105 | 0.01 |
ENST00000375994.2
|
MYLK2
|
myosin light chain kinase 2 |
| chr2_+_183580954 | 0.01 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr7_+_55177416 | 0.01 |
ENST00000450046.1
ENST00000454757.2 |
EGFR
|
epidermal growth factor receptor |
| chr5_+_178368186 | 0.01 |
ENST00000320129.3
ENST00000519564.1 |
ZNF454
|
zinc finger protein 454 |
| chr5_-_39074479 | 0.01 |
ENST00000514735.1
ENST00000296782.5 ENST00000357387.3 |
RICTOR
|
RPTOR independent companion of MTOR, complex 2 |
| chr10_-_33623310 | 0.01 |
ENST00000395995.1
ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1
|
neuropilin 1 |
| chr17_-_27053216 | 0.01 |
ENST00000292090.3
|
TLCD1
|
TLC domain containing 1 |
| chr6_+_42531798 | 0.01 |
ENST00000372903.2
ENST00000372899.1 ENST00000372901.1 |
UBR2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr2_+_204193101 | 0.01 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chr3_+_38017264 | 0.01 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr6_+_139135648 | 0.01 |
ENST00000541398.1
|
ECT2L
|
epithelial cell transforming sequence 2 oncogene-like |
| chr20_+_2633269 | 0.01 |
ENST00000445139.1
|
NOP56
|
NOP56 ribonucleoprotein |
| chr1_-_197036364 | 0.01 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr17_-_16118835 | 0.01 |
ENST00000582357.1
ENST00000436828.1 ENST00000411510.1 ENST00000268712.3 |
NCOR1
|
nuclear receptor corepressor 1 |
| chr5_+_178322893 | 0.01 |
ENST00000361362.2
ENST00000520660.1 ENST00000520805.1 |
ZFP2
|
ZFP2 zinc finger protein |
| chr19_-_37178284 | 0.01 |
ENST00000425254.2
ENST00000590952.1 ENST00000433232.1 |
AC074138.3
|
AC074138.3 |
| chr10_+_99205894 | 0.01 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr1_-_935519 | 0.01 |
ENST00000428771.2
|
HES4
|
hes family bHLH transcription factor 4 |
| chr16_-_57831676 | 0.01 |
ENST00000465878.2
ENST00000539578.1 ENST00000561524.1 |
KIFC3
|
kinesin family member C3 |
| chr20_+_30407151 | 0.01 |
ENST00000375985.4
|
MYLK2
|
myosin light chain kinase 2 |
| chr1_+_61547894 | 0.01 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr5_-_115872124 | 0.01 |
ENST00000515009.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr11_-_64647144 | 0.01 |
ENST00000359393.2
ENST00000433803.1 ENST00000411683.1 |
EHD1
|
EH-domain containing 1 |
| chrX_+_19362011 | 0.01 |
ENST00000379806.5
ENST00000545074.1 ENST00000540249.1 ENST00000423505.1 ENST00000417819.1 ENST00000422285.2 ENST00000355808.5 ENST00000379805.3 |
PDHA1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr9_-_104357277 | 0.01 |
ENST00000374806.1
|
PPP3R2
|
protein phosphatase 3, regulatory subunit B, beta |
| chr3_-_87325612 | 0.01 |
ENST00000561167.1
ENST00000560656.1 ENST00000344265.3 |
POU1F1
|
POU class 1 homeobox 1 |
| chr7_-_150924121 | 0.01 |
ENST00000441774.1
ENST00000222388.2 ENST00000287844.2 |
ABCF2
|
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr3_-_49967292 | 0.01 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chr19_+_39390587 | 0.01 |
ENST00000572515.1
ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr1_+_185126291 | 0.01 |
ENST00000367500.4
|
SWT1
|
SWT1 RNA endoribonuclease homolog (S. cerevisiae) |
| chr1_-_180471947 | 0.01 |
ENST00000367595.3
|
ACBD6
|
acyl-CoA binding domain containing 6 |
| chr12_+_106976678 | 0.01 |
ENST00000392842.1
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chr14_-_53417732 | 0.01 |
ENST00000399304.3
ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2
|
fermitin family member 2 |
| chr14_+_29234870 | 0.01 |
ENST00000382535.3
|
FOXG1
|
forkhead box G1 |
| chr5_-_124080203 | 0.01 |
ENST00000504926.1
|
ZNF608
|
zinc finger protein 608 |
| chr1_+_150980889 | 0.01 |
ENST00000450884.1
ENST00000271620.3 ENST00000271619.8 ENST00000368937.1 ENST00000431193.1 ENST00000368936.1 |
PRUNE
|
prune exopolyphosphatase |
| chr10_-_33623564 | 0.01 |
ENST00000374875.1
ENST00000374822.4 |
NRP1
|
neuropilin 1 |
| chr3_-_100712292 | 0.01 |
ENST00000495063.1
ENST00000530539.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr3_-_87325728 | 0.01 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr9_-_98269481 | 0.01 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chr15_-_70390191 | 0.01 |
ENST00000559191.1
|
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr10_-_33624002 | 0.00 |
ENST00000432372.2
|
NRP1
|
neuropilin 1 |
| chr20_+_44637526 | 0.00 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr1_+_150980989 | 0.00 |
ENST00000368935.1
|
PRUNE
|
prune exopolyphosphatase |
| chr4_-_164534657 | 0.00 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr2_-_170219037 | 0.00 |
ENST00000443831.1
|
LRP2
|
low density lipoprotein receptor-related protein 2 |
| chr15_-_33486897 | 0.00 |
ENST00000320930.7
|
FMN1
|
formin 1 |
| chrX_-_53713657 | 0.00 |
ENST00000262854.6
ENST00000218328.8 |
HUWE1
|
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
| chr1_+_8021954 | 0.00 |
ENST00000377491.1
ENST00000377488.1 |
PARK7
|
parkinson protein 7 |
| chr19_+_50706866 | 0.00 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr12_+_12938541 | 0.00 |
ENST00000356591.4
|
APOLD1
|
apolipoprotein L domain containing 1 |
| chr6_-_94129244 | 0.00 |
ENST00000369303.4
ENST00000369297.1 |
EPHA7
|
EPH receptor A7 |
| chr10_+_99205959 | 0.00 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr3_+_133524459 | 0.00 |
ENST00000484684.1
|
SRPRB
|
signal recognition particle receptor, B subunit |
| chrX_-_11369656 | 0.00 |
ENST00000413512.3
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr2_+_44001172 | 0.00 |
ENST00000260605.8
ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr2_+_38177620 | 0.00 |
ENST00000402091.3
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr2_+_48796120 | 0.00 |
ENST00000394754.1
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough |
| chr19_-_36870087 | 0.00 |
ENST00000270001.7
|
ZFP14
|
ZFP14 zinc finger protein |
| chr5_+_137514403 | 0.00 |
ENST00000513276.1
|
KIF20A
|
kinesin family member 20A |
| chr11_+_66384053 | 0.00 |
ENST00000310137.4
ENST00000393979.3 ENST00000409372.1 ENST00000443702.1 ENST00000409738.4 ENST00000514361.3 ENST00000503028.2 ENST00000412278.2 ENST00000500635.2 |
RBM14
RBM4
RBM14-RBM4
|
RNA binding motif protein 14 RNA binding motif protein 4 RBM14-RBM4 readthrough |
Network of associatons between targets according to the STRING database.
First level regulatory network of RBPJ
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0021699 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:1903567 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.4 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0002669 | regulation of tolerance induction dependent upon immune response(GO:0002652) positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 0.1 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.0 | 0.2 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.0 | GO:0032765 | positive regulation of mast cell cytokine production(GO:0032765) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |