Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for RARB
Z-value: 0.78
Transcription factors associated with RARB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARB
|
ENSG00000077092.14 | retinoic acid receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARB | hg19_v2_chr3_+_25469802_25469866 | 0.30 | 5.6e-01 | Click! |
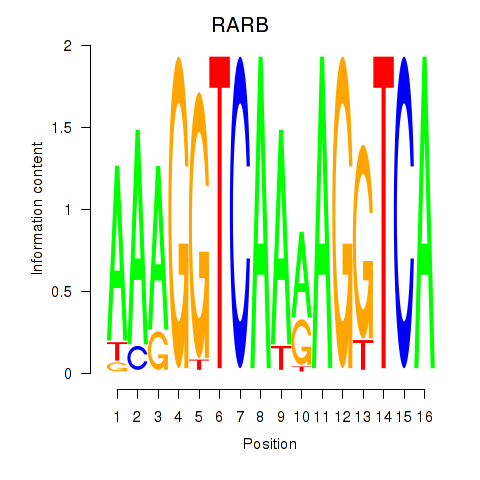
Activity profile of RARB motif
Sorted Z-values of RARB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_5914213 | 0.42 |
ENST00000222125.5
ENST00000452990.2 ENST00000588865.1 |
CAPS
|
calcyphosine |
| chr8_+_13424352 | 0.37 |
ENST00000297324.4
|
C8orf48
|
chromosome 8 open reading frame 48 |
| chr6_-_88038996 | 0.33 |
ENST00000525899.1
|
GJB7
|
gap junction protein, beta 7, 25kDa |
| chr6_-_150212029 | 0.30 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr2_+_132286754 | 0.29 |
ENST00000434330.1
|
CCDC74A
|
coiled-coil domain containing 74A |
| chr8_-_95274536 | 0.27 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr11_+_85566422 | 0.23 |
ENST00000342404.3
|
CCDC83
|
coiled-coil domain containing 83 |
| chr3_-_121448791 | 0.23 |
ENST00000489400.1
|
GOLGB1
|
golgin B1 |
| chr11_-_63381925 | 0.23 |
ENST00000415826.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr6_+_168434678 | 0.22 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr9_+_67977438 | 0.22 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr5_-_74162739 | 0.21 |
ENST00000513277.1
|
FAM169A
|
family with sequence similarity 169, member A |
| chr16_-_18923035 | 0.21 |
ENST00000563836.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr4_-_84406218 | 0.21 |
ENST00000515303.1
|
FAM175A
|
family with sequence similarity 175, member A |
| chr19_+_39279838 | 0.21 |
ENST00000314980.4
|
LGALS7B
|
lectin, galactoside-binding, soluble, 7B |
| chr12_+_4130143 | 0.21 |
ENST00000543206.1
|
RP11-320N7.2
|
RP11-320N7.2 |
| chrX_+_123095890 | 0.21 |
ENST00000435215.1
|
STAG2
|
stromal antigen 2 |
| chr12_+_57810198 | 0.20 |
ENST00000598001.1
|
AC126614.1
|
HCG1818482; Uncharacterized protein |
| chr16_+_2106134 | 0.20 |
ENST00000467949.1
|
TSC2
|
tuberous sclerosis 2 |
| chr19_-_58874112 | 0.18 |
ENST00000311044.3
ENST00000595763.1 ENST00000425453.3 |
ZNF497
|
zinc finger protein 497 |
| chr22_+_39493207 | 0.18 |
ENST00000348946.4
ENST00000442487.3 ENST00000421988.2 |
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr2_+_217082311 | 0.18 |
ENST00000597904.1
|
RP11-566E18.3
|
RP11-566E18.3 |
| chr12_+_57854274 | 0.17 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr12_-_113658826 | 0.17 |
ENST00000546692.1
|
IQCD
|
IQ motif containing D |
| chr19_+_42363917 | 0.17 |
ENST00000598742.1
|
RPS19
|
ribosomal protein S19 |
| chr2_-_208994548 | 0.16 |
ENST00000282141.3
|
CRYGC
|
crystallin, gamma C |
| chr7_-_144533074 | 0.16 |
ENST00000360057.3
ENST00000378099.3 |
TPK1
|
thiamin pyrophosphokinase 1 |
| chr1_+_1846519 | 0.16 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr8_-_36636676 | 0.16 |
ENST00000524132.1
ENST00000519451.1 |
RP11-962G15.1
|
RP11-962G15.1 |
| chr2_-_21266816 | 0.16 |
ENST00000399256.4
|
APOB
|
apolipoprotein B |
| chr17_+_41363854 | 0.16 |
ENST00000588693.1
ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A
|
transmembrane protein 106A |
| chr15_+_29211570 | 0.16 |
ENST00000558804.1
|
APBA2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr4_-_100140331 | 0.16 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr5_-_135290651 | 0.16 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chrX_-_128782722 | 0.16 |
ENST00000427399.1
|
APLN
|
apelin |
| chr19_-_22193731 | 0.15 |
ENST00000601773.1
ENST00000397126.4 ENST00000601993.1 ENST00000599916.1 |
ZNF208
|
zinc finger protein 208 |
| chr16_+_57673207 | 0.15 |
ENST00000564783.1
ENST00000564729.1 ENST00000565976.1 ENST00000566508.1 ENST00000544297.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr7_+_99425633 | 0.15 |
ENST00000354829.2
ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr11_-_8959758 | 0.15 |
ENST00000531618.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr19_-_58204128 | 0.15 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chrX_+_73164149 | 0.14 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr13_+_28519343 | 0.14 |
ENST00000381026.3
|
ATP5EP2
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr21_-_35284635 | 0.14 |
ENST00000429238.1
|
AP000304.12
|
AP000304.12 |
| chr10_-_103599591 | 0.14 |
ENST00000348850.5
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr6_+_49431073 | 0.13 |
ENST00000335783.3
|
CENPQ
|
centromere protein Q |
| chr19_-_22193706 | 0.13 |
ENST00000597040.1
|
ZNF208
|
zinc finger protein 208 |
| chr3_+_49297518 | 0.13 |
ENST00000440528.3
|
RP11-3B7.1
|
Uncharacterized protein |
| chr11_-_63381823 | 0.13 |
ENST00000323646.5
|
PLA2G16
|
phospholipase A2, group XVI |
| chr13_+_73302047 | 0.13 |
ENST00000377814.2
ENST00000377815.3 ENST00000390667.5 |
BORA
|
bora, aurora kinase A activator |
| chr1_-_110950564 | 0.13 |
ENST00000256644.4
|
LAMTOR5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr5_+_27472371 | 0.13 |
ENST00000512067.1
ENST00000510165.1 ENST00000505775.1 ENST00000514844.1 ENST00000503107.1 |
LINC01021
|
long intergenic non-protein coding RNA 1021 |
| chr10_-_81203972 | 0.13 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr15_-_31521567 | 0.12 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr20_-_44485835 | 0.12 |
ENST00000457981.1
ENST00000426915.1 ENST00000217455.4 |
ACOT8
|
acyl-CoA thioesterase 8 |
| chr6_-_133055896 | 0.12 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr4_-_48082192 | 0.12 |
ENST00000507351.1
|
TXK
|
TXK tyrosine kinase |
| chr11_-_111782696 | 0.12 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr17_+_48046671 | 0.12 |
ENST00000505318.2
|
DLX4
|
distal-less homeobox 4 |
| chr19_+_38880695 | 0.12 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr16_-_21875424 | 0.12 |
ENST00000541674.1
|
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr1_-_32687923 | 0.12 |
ENST00000309777.6
ENST00000344461.3 ENST00000373593.1 ENST00000545122.1 |
TMEM234
|
transmembrane protein 234 |
| chr1_+_52521928 | 0.12 |
ENST00000489308.2
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr5_-_180671172 | 0.12 |
ENST00000512805.1
|
GNB2L1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr19_+_50015870 | 0.12 |
ENST00000599701.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr16_+_2533020 | 0.11 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr7_-_5998714 | 0.11 |
ENST00000539903.1
|
RSPH10B
|
radial spoke head 10 homolog B (Chlamydomonas) |
| chr6_-_52859046 | 0.11 |
ENST00000457564.1
ENST00000541324.1 ENST00000370960.1 |
GSTA4
|
glutathione S-transferase alpha 4 |
| chr12_-_112443830 | 0.11 |
ENST00000550037.1
ENST00000549425.1 |
TMEM116
|
transmembrane protein 116 |
| chr1_-_150693318 | 0.11 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr10_-_8095412 | 0.11 |
ENST00000458727.1
ENST00000355358.1 |
RP11-379F12.3
GATA3-AS1
|
RP11-379F12.3 GATA3 antisense RNA 1 |
| chrX_+_123095860 | 0.11 |
ENST00000428941.1
|
STAG2
|
stromal antigen 2 |
| chr19_-_11688260 | 0.11 |
ENST00000590832.1
|
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr5_-_141392538 | 0.11 |
ENST00000503794.1
ENST00000510194.1 ENST00000504424.1 ENST00000513454.1 ENST00000458112.2 ENST00000542860.1 ENST00000503229.1 ENST00000500692.2 ENST00000311337.6 ENST00000504139.1 ENST00000505689.1 |
GNPDA1
|
glucosamine-6-phosphate deaminase 1 |
| chr9_-_104145795 | 0.11 |
ENST00000259407.2
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr17_+_73997419 | 0.11 |
ENST00000425876.2
|
CDK3
|
cyclin-dependent kinase 3 |
| chr14_+_104710541 | 0.11 |
ENST00000419115.1
|
C14orf144
|
chromosome 14 open reading frame 144 |
| chr1_+_35544968 | 0.11 |
ENST00000359858.4
ENST00000373330.1 |
ZMYM1
|
zinc finger, MYM-type 1 |
| chr12_+_113796347 | 0.11 |
ENST00000545182.2
ENST00000280800.3 |
PLBD2
|
phospholipase B domain containing 2 |
| chr1_+_160175166 | 0.10 |
ENST00000368077.1
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr11_-_798305 | 0.10 |
ENST00000531514.1
|
SLC25A22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr1_+_1215968 | 0.10 |
ENST00000338555.2
|
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chr12_+_56075330 | 0.10 |
ENST00000394252.3
|
METTL7B
|
methyltransferase like 7B |
| chr8_+_39770803 | 0.10 |
ENST00000518237.1
|
IDO1
|
indoleamine 2,3-dioxygenase 1 |
| chr17_+_38375528 | 0.10 |
ENST00000583268.1
|
WIPF2
|
WAS/WASL interacting protein family, member 2 |
| chr1_-_111506562 | 0.10 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr4_-_69536346 | 0.10 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr19_+_21688366 | 0.10 |
ENST00000358491.4
ENST00000597078.1 |
ZNF429
|
zinc finger protein 429 |
| chr1_-_150979333 | 0.10 |
ENST00000312210.5
|
FAM63A
|
family with sequence similarity 63, member A |
| chr12_+_93963590 | 0.10 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr15_-_54051831 | 0.10 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr4_+_56719782 | 0.10 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr1_-_110950255 | 0.09 |
ENST00000483260.1
ENST00000474861.2 ENST00000602318.1 |
LAMTOR5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr17_+_28884130 | 0.09 |
ENST00000580161.1
|
TBC1D29
|
TBC1 domain family, member 29 |
| chrX_+_23682379 | 0.09 |
ENST00000379349.1
|
PRDX4
|
peroxiredoxin 4 |
| chr7_-_99381798 | 0.09 |
ENST00000415003.1
ENST00000354593.2 |
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr1_-_52521831 | 0.09 |
ENST00000371626.4
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr10_+_6779326 | 0.09 |
ENST00000417112.1
|
RP11-554I8.2
|
RP11-554I8.2 |
| chr5_-_74162605 | 0.09 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr3_+_99986036 | 0.09 |
ENST00000471098.1
|
TBC1D23
|
TBC1 domain family, member 23 |
| chr10_-_43762329 | 0.08 |
ENST00000395810.1
|
RASGEF1A
|
RasGEF domain family, member 1A |
| chr16_-_20587599 | 0.08 |
ENST00000566384.1
ENST00000565232.1 ENST00000567001.1 ENST00000565322.1 ENST00000569344.1 ENST00000329697.6 ENST00000414188.2 ENST00000568882.1 |
ACSM2B
|
acyl-CoA synthetase medium-chain family member 2B |
| chr6_-_150217195 | 0.08 |
ENST00000532335.1
|
RAET1E
|
retinoic acid early transcript 1E |
| chr1_+_52521797 | 0.08 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr5_-_157161727 | 0.08 |
ENST00000599823.1
|
AC026407.1
|
Uncharacterized protein |
| chr2_+_7118755 | 0.08 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr3_-_49837254 | 0.08 |
ENST00000412678.2
ENST00000343366.4 ENST00000487256.1 |
CDHR4
|
cadherin-related family member 4 |
| chr12_+_6881678 | 0.07 |
ENST00000441671.2
ENST00000203629.2 |
LAG3
|
lymphocyte-activation gene 3 |
| chr19_-_51017127 | 0.07 |
ENST00000389208.4
|
ASPDH
|
aspartate dehydrogenase domain containing |
| chr1_-_150693305 | 0.07 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chr10_-_65028817 | 0.07 |
ENST00000542921.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr1_+_120049809 | 0.07 |
ENST00000531340.1
|
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr11_-_116708302 | 0.07 |
ENST00000375320.1
ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1
|
apolipoprotein A-I |
| chr3_+_119316689 | 0.07 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr1_+_1215816 | 0.07 |
ENST00000379116.5
|
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chr6_+_2988199 | 0.07 |
ENST00000450238.1
ENST00000445000.1 ENST00000426637.1 |
LINC01011
NQO2
|
long intergenic non-protein coding RNA 1011 NAD(P)H dehydrogenase, quinone 2 |
| chrX_-_119709637 | 0.07 |
ENST00000404115.3
|
CUL4B
|
cullin 4B |
| chr22_-_43036607 | 0.07 |
ENST00000505920.1
|
ATP5L2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr1_-_86861660 | 0.06 |
ENST00000486215.1
|
ODF2L
|
outer dense fiber of sperm tails 2-like |
| chr17_-_79784533 | 0.06 |
ENST00000457257.1
ENST00000576730.1 |
AC174470.1
FAM195B
|
AC174470.1 family with sequence similarity 195, member B |
| chr2_+_114737146 | 0.06 |
ENST00000412690.1
|
AC010982.1
|
AC010982.1 |
| chr5_+_140174429 | 0.06 |
ENST00000520672.2
ENST00000378132.1 ENST00000526136.1 |
PCDHA2
|
protocadherin alpha 2 |
| chr6_+_139456226 | 0.06 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr9_-_100459639 | 0.06 |
ENST00000375128.4
|
XPA
|
xeroderma pigmentosum, complementation group A |
| chr19_-_8567505 | 0.06 |
ENST00000600262.1
|
PRAM1
|
PML-RARA regulated adaptor molecule 1 |
| chr1_+_78245303 | 0.06 |
ENST00000370791.3
ENST00000443751.2 |
FAM73A
|
family with sequence similarity 73, member A |
| chrM_+_3299 | 0.06 |
ENST00000361390.2
|
MT-ND1
|
mitochondrially encoded NADH dehydrogenase 1 |
| chr6_+_31554612 | 0.06 |
ENST00000211921.7
|
LST1
|
leukocyte specific transcript 1 |
| chr22_-_37976082 | 0.06 |
ENST00000215886.4
|
LGALS2
|
lectin, galactoside-binding, soluble, 2 |
| chr19_-_33360647 | 0.06 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr11_-_111782484 | 0.06 |
ENST00000533971.1
|
CRYAB
|
crystallin, alpha B |
| chr2_+_74361599 | 0.06 |
ENST00000401851.1
|
MGC10955
|
MGC10955 |
| chr11_+_63870660 | 0.06 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr16_-_4852915 | 0.06 |
ENST00000322048.7
|
ROGDI
|
rogdi homolog (Drosophila) |
| chr12_-_6756609 | 0.05 |
ENST00000229243.2
|
ACRBP
|
acrosin binding protein |
| chr4_+_124317940 | 0.05 |
ENST00000505319.1
ENST00000339241.1 |
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr17_-_41277317 | 0.05 |
ENST00000497488.1
ENST00000489037.1 ENST00000470026.1 ENST00000586385.1 ENST00000591534.1 ENST00000591849.1 |
BRCA1
|
breast cancer 1, early onset |
| chr3_+_119316721 | 0.05 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr12_-_323248 | 0.05 |
ENST00000535347.1
ENST00000536824.1 |
SLC6A12
|
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr7_-_44180884 | 0.05 |
ENST00000458240.1
ENST00000223364.3 |
MYL7
|
myosin, light chain 7, regulatory |
| chr1_+_120049826 | 0.05 |
ENST00000369413.3
ENST00000235547.6 ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr22_+_39493268 | 0.05 |
ENST00000401756.1
|
APOBEC3H
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
| chr11_-_1587166 | 0.05 |
ENST00000331588.4
|
DUSP8
|
dual specificity phosphatase 8 |
| chr4_-_168155300 | 0.05 |
ENST00000541637.1
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr2_-_97405775 | 0.05 |
ENST00000264963.4
ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L
|
lectin, mannose-binding 2-like |
| chr4_+_8594435 | 0.05 |
ENST00000382480.2
|
CPZ
|
carboxypeptidase Z |
| chr3_+_184058125 | 0.05 |
ENST00000310585.4
|
FAM131A
|
family with sequence similarity 131, member A |
| chr9_+_35909478 | 0.05 |
ENST00000443779.1
|
LINC00961
|
long intergenic non-protein coding RNA 961 |
| chr10_-_65028938 | 0.05 |
ENST00000402544.1
|
JMJD1C
|
jumonji domain containing 1C |
| chr2_-_176866978 | 0.05 |
ENST00000392540.2
ENST00000409660.1 ENST00000544803.1 ENST00000272748.4 |
KIAA1715
|
KIAA1715 |
| chr8_+_74903580 | 0.05 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr4_+_8594477 | 0.05 |
ENST00000315782.6
|
CPZ
|
carboxypeptidase Z |
| chr6_+_76311568 | 0.05 |
ENST00000370014.3
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr3_+_14862939 | 0.05 |
ENST00000457774.1
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5 |
| chr17_-_74287333 | 0.05 |
ENST00000447564.2
|
QRICH2
|
glutamine rich 2 |
| chr7_+_20686946 | 0.05 |
ENST00000443026.2
ENST00000406935.1 |
ABCB5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr18_-_70210764 | 0.05 |
ENST00000585159.1
ENST00000584764.1 |
CBLN2
|
cerebellin 2 precursor |
| chr17_-_44657017 | 0.05 |
ENST00000573185.1
ENST00000570550.1 ENST00000445552.2 ENST00000336125.5 ENST00000329240.4 ENST00000337845.7 |
ARL17A
|
ADP-ribosylation factor-like 17A |
| chr1_+_174669653 | 0.05 |
ENST00000325589.5
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr20_-_43729750 | 0.05 |
ENST00000537075.1
ENST00000306117.1 |
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
| chr1_+_155278539 | 0.05 |
ENST00000447866.1
|
FDPS
|
farnesyl diphosphate synthase |
| chr16_+_2255710 | 0.04 |
ENST00000397124.1
ENST00000565250.1 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr1_+_9005917 | 0.04 |
ENST00000549778.1
ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6
|
carbonic anhydrase VI |
| chr4_+_8594364 | 0.04 |
ENST00000360986.4
|
CPZ
|
carboxypeptidase Z |
| chr18_-_32924372 | 0.04 |
ENST00000261332.6
ENST00000399061.3 |
ZNF24
|
zinc finger protein 24 |
| chr7_-_141401951 | 0.04 |
ENST00000536163.1
|
KIAA1147
|
KIAA1147 |
| chr1_+_160175117 | 0.04 |
ENST00000360472.4
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chrX_+_73164167 | 0.04 |
ENST00000414209.1
ENST00000602895.1 ENST00000453317.1 ENST00000602546.1 ENST00000602985.1 ENST00000415215.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chr6_-_24667180 | 0.04 |
ENST00000545995.1
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr6_-_24667232 | 0.04 |
ENST00000378198.4
|
TDP2
|
tyrosyl-DNA phosphodiesterase 2 |
| chr10_-_70287231 | 0.04 |
ENST00000609923.1
|
SLC25A16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr11_-_506316 | 0.04 |
ENST00000532055.1
ENST00000531540.1 |
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr6_-_71012773 | 0.04 |
ENST00000370496.3
ENST00000357250.6 |
COL9A1
|
collagen, type IX, alpha 1 |
| chr4_+_70146217 | 0.04 |
ENST00000335568.5
ENST00000511240.1 |
UGT2B28
|
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chrX_-_53461288 | 0.04 |
ENST00000375298.4
ENST00000375304.5 |
HSD17B10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr11_-_95522907 | 0.04 |
ENST00000358780.5
ENST00000542135.1 |
FAM76B
|
family with sequence similarity 76, member B |
| chr1_+_248020481 | 0.04 |
ENST00000366481.3
|
TRIM58
|
tripartite motif containing 58 |
| chrX_+_102024075 | 0.04 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr19_-_17559376 | 0.03 |
ENST00000341130.5
|
TMEM221
|
transmembrane protein 221 |
| chr6_-_49430886 | 0.03 |
ENST00000274813.3
|
MUT
|
methylmalonyl CoA mutase |
| chr1_-_204135450 | 0.03 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr2_+_119981384 | 0.03 |
ENST00000393108.2
ENST00000354888.5 ENST00000450943.2 ENST00000393110.2 ENST00000393106.2 ENST00000409811.1 ENST00000393107.2 |
STEAP3
|
STEAP family member 3, metalloreductase |
| chr18_+_19192228 | 0.03 |
ENST00000300413.5
ENST00000579618.1 ENST00000582475.1 |
SNRPD1
|
small nuclear ribonucleoprotein D1 polypeptide 16kDa |
| chr19_-_37406923 | 0.03 |
ENST00000520965.1
|
ZNF829
|
zinc finger protein 829 |
| chr12_+_131438443 | 0.03 |
ENST00000261654.5
|
GPR133
|
G protein-coupled receptor 133 |
| chr19_+_1491144 | 0.03 |
ENST00000233596.3
|
REEP6
|
receptor accessory protein 6 |
| chr18_-_19283649 | 0.03 |
ENST00000584464.1
ENST00000578270.1 |
ABHD3
|
abhydrolase domain containing 3 |
| chr10_-_33625154 | 0.03 |
ENST00000265371.4
|
NRP1
|
neuropilin 1 |
| chr1_-_23521222 | 0.03 |
ENST00000374619.1
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chrX_+_148769880 | 0.03 |
ENST00000412632.2
|
MAGEA11
|
melanoma antigen family A, 11 |
| chr5_-_99870932 | 0.03 |
ENST00000504833.1
|
CTD-2001C12.1
|
CTD-2001C12.1 |
| chr11_+_95523621 | 0.03 |
ENST00000325542.5
ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57
|
centrosomal protein 57kDa |
| chr12_+_49658855 | 0.03 |
ENST00000549183.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr11_-_116663127 | 0.03 |
ENST00000433069.1
ENST00000542499.1 |
APOA5
|
apolipoprotein A-V |
| chr17_-_62097904 | 0.03 |
ENST00000583366.1
|
ICAM2
|
intercellular adhesion molecule 2 |
| chr6_+_76311636 | 0.03 |
ENST00000327284.8
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr11_+_121447469 | 0.03 |
ENST00000532694.1
ENST00000534286.1 |
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr19_+_49468558 | 0.03 |
ENST00000331825.6
|
FTL
|
ferritin, light polypeptide |
| chr6_+_31637944 | 0.03 |
ENST00000375864.4
|
LY6G5B
|
lymphocyte antigen 6 complex, locus G5B |
| chr5_-_135290705 | 0.03 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr19_+_21324827 | 0.03 |
ENST00000600692.1
ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431
|
zinc finger protein 431 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RARB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.1 | 0.2 | GO:0060629 | regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.2 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.2 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.2 | GO:0060266 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.0 | 0.1 | GO:0006043 | glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.2 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.2 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0060332 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.0 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.1 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.1 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0036317 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0034191 | apolipoprotein A-I receptor binding(GO:0034191) phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |