Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
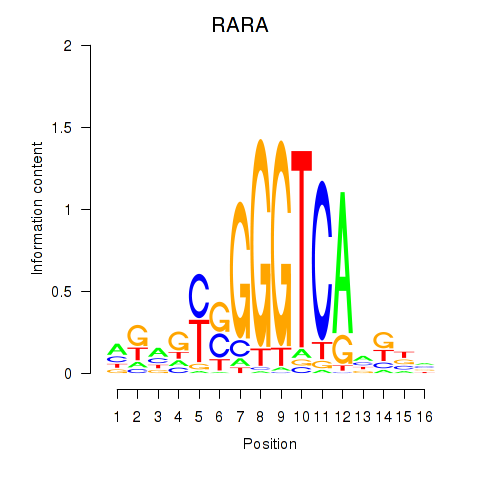
Results for RARA
Z-value: 1.26
Transcription factors associated with RARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARA
|
ENSG00000131759.13 | retinoic acid receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARA | hg19_v2_chr17_+_38474489_38474548 | -0.72 | 1.1e-01 | Click! |
Activity profile of RARA motif
Sorted Z-values of RARA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_24641062 | 1.09 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr22_-_50970566 | 0.99 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr8_-_101348408 | 0.99 |
ENST00000519527.1
ENST00000522369.1 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr12_-_44152551 | 0.98 |
ENST00000416848.2
ENST00000550784.1 ENST00000547156.1 ENST00000549868.1 ENST00000553166.1 ENST00000551923.1 ENST00000431332.3 ENST00000344862.5 |
PUS7L
|
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
| chr1_-_93426998 | 0.85 |
ENST00000370310.4
|
FAM69A
|
family with sequence similarity 69, member A |
| chr22_-_50970506 | 0.84 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr3_-_88108192 | 0.80 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr8_+_104152922 | 0.79 |
ENST00000309982.5
ENST00000438105.2 ENST00000297574.6 |
BAALC
|
brain and acute leukemia, cytoplasmic |
| chr9_+_139873264 | 0.79 |
ENST00000446677.1
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr7_+_130794878 | 0.76 |
ENST00000416992.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr18_+_44526786 | 0.76 |
ENST00000245121.5
ENST00000356157.7 |
KATNAL2
|
katanin p60 subunit A-like 2 |
| chr14_-_45603657 | 0.72 |
ENST00000396062.3
|
FKBP3
|
FK506 binding protein 3, 25kDa |
| chr6_-_35888905 | 0.69 |
ENST00000510290.1
ENST00000423325.2 ENST00000373822.1 |
SRPK1
|
SRSF protein kinase 1 |
| chr9_-_86432547 | 0.68 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr12_-_123459105 | 0.65 |
ENST00000543935.1
|
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr7_-_105752971 | 0.63 |
ENST00000011473.2
|
SYPL1
|
synaptophysin-like 1 |
| chr14_-_54955376 | 0.62 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr19_-_19739321 | 0.62 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr17_-_16557128 | 0.62 |
ENST00000423860.2
ENST00000311331.7 ENST00000583766.1 |
ZNF624
|
zinc finger protein 624 |
| chr10_+_60145155 | 0.59 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr1_-_109203685 | 0.59 |
ENST00000402983.1
ENST00000420055.1 |
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chrX_-_37706815 | 0.58 |
ENST00000378578.4
|
DYNLT3
|
dynein, light chain, Tctex-type 3 |
| chr19_-_6720686 | 0.58 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr10_+_115438920 | 0.58 |
ENST00000429617.1
ENST00000369331.4 |
CASP7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr14_+_54976603 | 0.57 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr6_+_12008986 | 0.56 |
ENST00000491710.1
|
HIVEP1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chrX_-_102319092 | 0.56 |
ENST00000372728.3
|
BEX1
|
brain expressed, X-linked 1 |
| chr4_+_186392597 | 0.55 |
ENST00000512874.1
|
RP11-279O9.4
|
Uncharacterized protein |
| chr1_+_219347203 | 0.55 |
ENST00000366927.3
|
LYPLAL1
|
lysophospholipase-like 1 |
| chr5_-_114961858 | 0.55 |
ENST00000282382.4
ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2
TMED7
TICAM2
|
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr13_-_52026730 | 0.55 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr15_+_76135622 | 0.55 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr2_+_189157536 | 0.55 |
ENST00000409580.1
ENST00000409637.3 |
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr4_-_186125077 | 0.54 |
ENST00000458385.2
ENST00000514798.1 ENST00000296775.6 |
KIAA1430
|
KIAA1430 |
| chr2_-_113191096 | 0.54 |
ENST00000496537.1
ENST00000330575.5 ENST00000302558.3 |
RGPD8
|
RANBP2-like and GRIP domain containing 8 |
| chr3_-_101232019 | 0.54 |
ENST00000394095.2
ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7
|
SUMO1/sentrin specific peptidase 7 |
| chr10_+_62538248 | 0.54 |
ENST00000448257.2
|
CDK1
|
cyclin-dependent kinase 1 |
| chr1_-_85156417 | 0.53 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr4_+_107236692 | 0.53 |
ENST00000510207.1
|
AIMP1
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
| chr4_-_159592996 | 0.53 |
ENST00000508457.1
|
C4orf46
|
chromosome 4 open reading frame 46 |
| chr19_-_37958086 | 0.52 |
ENST00000592490.1
ENST00000392149.2 |
ZNF569
|
zinc finger protein 569 |
| chr4_+_25915896 | 0.52 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr4_+_80584903 | 0.52 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
| chr12_+_67663056 | 0.52 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr5_-_135290651 | 0.51 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr5_-_55290773 | 0.51 |
ENST00000502326.3
ENST00000381298.2 |
IL6ST
|
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr15_+_33010175 | 0.51 |
ENST00000300177.4
ENST00000560677.1 ENST00000560830.1 |
GREM1
|
gremlin 1, DAN family BMP antagonist |
| chr7_+_106810165 | 0.51 |
ENST00000468401.1
ENST00000497535.1 ENST00000485846.1 |
HBP1
|
HMG-box transcription factor 1 |
| chr7_-_148725544 | 0.51 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr4_+_159593418 | 0.51 |
ENST00000507475.1
ENST00000307738.5 |
ETFDH
|
electron-transferring-flavoprotein dehydrogenase |
| chr11_-_18610214 | 0.50 |
ENST00000300038.7
ENST00000396197.3 ENST00000320750.6 |
UEVLD
|
UEV and lactate/malate dehyrogenase domains |
| chr6_+_108616243 | 0.50 |
ENST00000421954.1
|
LACE1
|
lactation elevated 1 |
| chr10_-_126849626 | 0.50 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr1_+_76251912 | 0.49 |
ENST00000370826.3
|
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr13_-_48575401 | 0.49 |
ENST00000433022.1
ENST00000544100.1 |
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr5_+_65440032 | 0.49 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr4_+_146019421 | 0.49 |
ENST00000502586.1
|
ABCE1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr9_-_72374848 | 0.48 |
ENST00000377200.5
ENST00000340434.4 ENST00000472967.2 |
PTAR1
|
protein prenyltransferase alpha subunit repeat containing 1 |
| chr8_-_103136481 | 0.48 |
ENST00000524209.1
ENST00000517822.1 ENST00000523923.1 ENST00000521599.1 ENST00000521964.1 ENST00000311028.3 ENST00000518166.1 |
NCALD
|
neurocalcin delta |
| chr13_-_48575443 | 0.48 |
ENST00000378654.3
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr19_-_821931 | 0.48 |
ENST00000359894.2
ENST00000520876.3 ENST00000519502.1 |
LPPR3
|
hsa-mir-3187 |
| chr11_+_32605350 | 0.48 |
ENST00000531120.1
ENST00000524896.1 ENST00000323213.5 |
EIF3M
|
eukaryotic translation initiation factor 3, subunit M |
| chr10_+_62538089 | 0.48 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr1_+_222988406 | 0.48 |
ENST00000448808.1
ENST00000457636.1 ENST00000439440.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr8_-_74659068 | 0.48 |
ENST00000523558.1
ENST00000521210.1 ENST00000355780.5 ENST00000524104.1 ENST00000521736.1 ENST00000521447.1 ENST00000517542.1 ENST00000521451.1 ENST00000521419.1 ENST00000518502.1 ENST00000524300.1 |
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr1_-_200379180 | 0.47 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr1_-_111506562 | 0.47 |
ENST00000485275.2
ENST00000369763.4 |
LRIF1
|
ligand dependent nuclear receptor interacting factor 1 |
| chr18_-_812517 | 0.47 |
ENST00000584307.1
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr14_-_71107921 | 0.46 |
ENST00000553982.1
ENST00000500016.1 |
CTD-2540L5.5
CTD-2540L5.6
|
CTD-2540L5.5 CTD-2540L5.6 |
| chr11_+_105948216 | 0.46 |
ENST00000278618.4
|
AASDHPPT
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr15_+_23255242 | 0.46 |
ENST00000450802.3
|
GOLGA8I
|
golgin A8 family, member I |
| chr9_+_114393581 | 0.46 |
ENST00000313525.3
|
DNAJC25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr2_-_219433014 | 0.46 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr6_+_139349903 | 0.45 |
ENST00000461027.1
|
ABRACL
|
ABRA C-terminal like |
| chr10_+_60144782 | 0.45 |
ENST00000487519.1
|
TFAM
|
transcription factor A, mitochondrial |
| chr6_+_111303218 | 0.44 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae) |
| chr7_-_105752651 | 0.44 |
ENST00000470347.1
ENST00000455385.2 |
SYPL1
|
synaptophysin-like 1 |
| chr11_+_33037401 | 0.43 |
ENST00000241051.3
|
DEPDC7
|
DEP domain containing 7 |
| chr4_-_184580304 | 0.43 |
ENST00000510968.1
ENST00000512740.1 ENST00000327570.9 |
RWDD4
|
RWD domain containing 4 |
| chr13_+_73356197 | 0.43 |
ENST00000326291.6
|
PIBF1
|
progesterone immunomodulatory binding factor 1 |
| chr1_-_109203648 | 0.43 |
ENST00000370031.1
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr1_-_59165763 | 0.42 |
ENST00000472487.1
|
MYSM1
|
Myb-like, SWIRM and MPN domains 1 |
| chr5_-_87564620 | 0.41 |
ENST00000506536.1
ENST00000512429.1 ENST00000514135.1 ENST00000296595.6 ENST00000509387.1 |
TMEM161B
|
transmembrane protein 161B |
| chr9_+_79792410 | 0.41 |
ENST00000357409.5
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr11_-_17410869 | 0.41 |
ENST00000528731.1
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr16_-_14724131 | 0.41 |
ENST00000341484.7
|
PARN
|
poly(A)-specific ribonuclease |
| chr7_+_130794846 | 0.41 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr11_-_102323489 | 0.41 |
ENST00000361236.3
|
TMEM123
|
transmembrane protein 123 |
| chr22_-_29137771 | 0.41 |
ENST00000439200.1
ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2
|
checkpoint kinase 2 |
| chr14_+_103566665 | 0.41 |
ENST00000559116.1
|
EXOC3L4
|
exocyst complex component 3-like 4 |
| chr19_+_35168633 | 0.40 |
ENST00000505365.2
|
ZNF302
|
zinc finger protein 302 |
| chr6_-_57086200 | 0.40 |
ENST00000468148.1
|
RAB23
|
RAB23, member RAS oncogene family |
| chr19_-_44331332 | 0.40 |
ENST00000602179.1
|
LYPD5
|
LY6/PLAUR domain containing 5 |
| chr8_+_87354945 | 0.40 |
ENST00000517970.1
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr1_+_174969262 | 0.39 |
ENST00000406752.1
ENST00000405362.1 |
CACYBP
|
calcyclin binding protein |
| chr18_-_812231 | 0.39 |
ENST00000314574.4
|
YES1
|
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr1_-_113498616 | 0.39 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr4_-_146019287 | 0.39 |
ENST00000502847.1
ENST00000513054.1 |
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr5_+_131892815 | 0.39 |
ENST00000453394.1
|
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr22_-_38349552 | 0.39 |
ENST00000422191.1
ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23
|
chromosome 22 open reading frame 23 |
| chr4_-_146019693 | 0.39 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr11_-_18610275 | 0.38 |
ENST00000543987.1
|
UEVLD
|
UEV and lactate/malate dehyrogenase domains |
| chr6_+_151358048 | 0.38 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr22_+_36649170 | 0.38 |
ENST00000438034.1
ENST00000427990.1 ENST00000347595.7 ENST00000397279.4 ENST00000433768.1 ENST00000440669.2 |
APOL1
|
apolipoprotein L, 1 |
| chr1_-_156675368 | 0.38 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr6_-_127664475 | 0.38 |
ENST00000474289.2
ENST00000534442.1 ENST00000368289.2 ENST00000525745.1 ENST00000430841.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr2_+_157291953 | 0.38 |
ENST00000310454.6
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr6_+_134273300 | 0.38 |
ENST00000416965.1
|
TBPL1
|
TBP-like 1 |
| chr1_-_109203997 | 0.38 |
ENST00000370032.5
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr7_-_108166505 | 0.38 |
ENST00000426128.2
ENST00000427008.1 ENST00000388728.5 ENST00000257694.8 ENST00000422087.1 ENST00000453144.1 ENST00000436062.1 |
PNPLA8
|
patatin-like phospholipase domain containing 8 |
| chr11_+_57310114 | 0.37 |
ENST00000527972.1
ENST00000399154.2 |
SMTNL1
|
smoothelin-like 1 |
| chr1_+_222988363 | 0.37 |
ENST00000450784.1
ENST00000426045.1 ENST00000457955.1 ENST00000444858.1 ENST00000435378.1 ENST00000441676.1 |
RP11-452F19.3
|
RP11-452F19.3 |
| chr2_+_189157498 | 0.37 |
ENST00000359135.3
|
GULP1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr6_+_96025341 | 0.37 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr9_-_88969303 | 0.37 |
ENST00000277141.6
ENST00000375963.3 |
ZCCHC6
|
zinc finger, CCHC domain containing 6 |
| chr18_+_12947981 | 0.37 |
ENST00000262124.11
|
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr2_-_113594279 | 0.37 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr13_+_53030107 | 0.37 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr1_-_222763101 | 0.36 |
ENST00000391883.2
ENST00000366890.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr17_-_80291818 | 0.36 |
ENST00000269389.3
ENST00000581691.1 |
SECTM1
|
secreted and transmembrane 1 |
| chr18_-_6929797 | 0.36 |
ENST00000581725.1
ENST00000583316.1 |
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr11_+_114310237 | 0.36 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr1_-_161008697 | 0.36 |
ENST00000318289.10
ENST00000368023.3 ENST00000368024.1 ENST00000423014.2 |
TSTD1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr17_-_6554747 | 0.36 |
ENST00000574128.1
|
MED31
|
mediator complex subunit 31 |
| chr12_+_133613878 | 0.36 |
ENST00000392319.2
ENST00000543758.1 |
ZNF84
|
zinc finger protein 84 |
| chr9_+_114393634 | 0.36 |
ENST00000556107.1
ENST00000374294.3 |
DNAJC25
DNAJC25-GNG10
|
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr1_+_91966384 | 0.36 |
ENST00000430031.2
ENST00000234626.6 |
CDC7
|
cell division cycle 7 |
| chr1_+_109289279 | 0.36 |
ENST00000370008.3
|
STXBP3
|
syntaxin binding protein 3 |
| chr9_-_88969339 | 0.36 |
ENST00000375960.2
ENST00000375961.2 |
ZCCHC6
|
zinc finger, CCHC domain containing 6 |
| chr9_+_86595626 | 0.35 |
ENST00000445877.1
ENST00000325875.3 |
RMI1
|
RecQ mediated genome instability 1 |
| chr2_+_47630108 | 0.35 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr5_-_171433548 | 0.35 |
ENST00000517395.1
|
FBXW11
|
F-box and WD repeat domain containing 11 |
| chr2_+_47630255 | 0.35 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr5_-_43313574 | 0.34 |
ENST00000325110.6
ENST00000433297.2 |
HMGCS1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr7_+_149416439 | 0.34 |
ENST00000497895.1
|
KRBA1
|
KRAB-A domain containing 1 |
| chrX_+_69501943 | 0.34 |
ENST00000509895.1
ENST00000374473.2 ENST00000276066.4 |
RAB41
|
RAB41, member RAS oncogene family |
| chr10_+_49514698 | 0.34 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr10_+_1102303 | 0.34 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr14_+_69726968 | 0.33 |
ENST00000553669.1
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr13_+_53029564 | 0.33 |
ENST00000468284.1
ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr1_+_145439306 | 0.33 |
ENST00000425134.1
|
TXNIP
|
thioredoxin interacting protein |
| chr5_+_112312454 | 0.33 |
ENST00000543319.1
|
DCP2
|
decapping mRNA 2 |
| chr1_+_203765437 | 0.33 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr13_-_73356009 | 0.33 |
ENST00000377780.4
ENST00000377767.4 |
DIS3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr15_+_80351977 | 0.33 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr10_-_112678692 | 0.32 |
ENST00000605742.1
|
BBIP1
|
BBSome interacting protein 1 |
| chr18_+_12948000 | 0.32 |
ENST00000585730.1
ENST00000399892.2 ENST00000589446.1 ENST00000587761.1 |
SEH1L
|
SEH1-like (S. cerevisiae) |
| chr5_+_118691008 | 0.32 |
ENST00000504642.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chrX_+_48534985 | 0.32 |
ENST00000450772.1
|
WAS
|
Wiskott-Aldrich syndrome |
| chr20_-_25604811 | 0.32 |
ENST00000304788.3
|
NANP
|
N-acetylneuraminic acid phosphatase |
| chr7_-_100239132 | 0.32 |
ENST00000223051.3
ENST00000431692.1 |
TFR2
|
transferrin receptor 2 |
| chr5_+_31532373 | 0.32 |
ENST00000325366.9
ENST00000355907.3 ENST00000507818.2 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr9_-_135230336 | 0.32 |
ENST00000224140.5
ENST00000372169.2 ENST00000393220.1 |
SETX
|
senataxin |
| chr3_-_185641681 | 0.32 |
ENST00000259043.7
|
TRA2B
|
transformer 2 beta homolog (Drosophila) |
| chr11_+_9482551 | 0.32 |
ENST00000438144.2
ENST00000526657.1 ENST00000299606.2 ENST00000534265.1 ENST00000412390.2 |
ZNF143
|
zinc finger protein 143 |
| chr1_+_222791417 | 0.31 |
ENST00000344922.5
ENST00000344441.6 ENST00000344507.1 |
MIA3
|
melanoma inhibitory activity family, member 3 |
| chr4_-_107237340 | 0.31 |
ENST00000394706.3
|
TBCK
|
TBC1 domain containing kinase |
| chr6_-_110500905 | 0.31 |
ENST00000392587.2
|
WASF1
|
WAS protein family, member 1 |
| chr9_-_125027079 | 0.31 |
ENST00000417201.3
|
RBM18
|
RNA binding motif protein 18 |
| chr7_+_30068260 | 0.30 |
ENST00000440706.2
|
PLEKHA8
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| chr1_-_222763240 | 0.30 |
ENST00000352967.4
ENST00000391882.1 ENST00000543857.1 |
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr21_-_34144157 | 0.30 |
ENST00000331923.4
|
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr12_+_7072354 | 0.30 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr1_-_222763214 | 0.30 |
ENST00000350027.4
|
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chr5_+_118690466 | 0.30 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr6_-_127664736 | 0.29 |
ENST00000368291.2
ENST00000309620.9 ENST00000454859.3 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr10_+_43633914 | 0.29 |
ENST00000374466.3
ENST00000374464.1 |
CSGALNACT2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr18_-_6929829 | 0.29 |
ENST00000583840.1
ENST00000581571.1 ENST00000578497.1 ENST00000579012.1 |
LINC00668
|
long intergenic non-protein coding RNA 668 |
| chr6_-_41040195 | 0.29 |
ENST00000463088.1
ENST00000469104.1 ENST00000486443.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr17_+_10048831 | 0.29 |
ENST00000581040.1
|
AC000003.2
|
CDNA FLJ25865 fis, clone CBR01927 |
| chr2_-_208030886 | 0.28 |
ENST00000426163.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr11_-_33795893 | 0.28 |
ENST00000526785.1
ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3
|
F-box protein 3 |
| chr14_+_24605389 | 0.28 |
ENST00000382708.3
ENST00000561435.1 |
PSME1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr6_-_127664683 | 0.28 |
ENST00000528402.1
ENST00000454591.2 |
ECHDC1
|
enoyl CoA hydratase domain containing 1 |
| chr13_+_43597269 | 0.28 |
ENST00000379221.2
|
DNAJC15
|
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr21_+_35445827 | 0.28 |
ENST00000608209.1
ENST00000381151.3 |
SLC5A3
SLC5A3
|
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr4_-_159593179 | 0.28 |
ENST00000379205.4
|
C4orf46
|
chromosome 4 open reading frame 46 |
| chr14_+_35761540 | 0.28 |
ENST00000261479.4
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr10_-_112678904 | 0.28 |
ENST00000423273.1
ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1
|
BBSome interacting protein 1 |
| chr3_-_123680246 | 0.28 |
ENST00000488653.2
|
CCDC14
|
coiled-coil domain containing 14 |
| chr4_+_184580420 | 0.27 |
ENST00000334690.6
ENST00000357207.4 |
TRAPPC11
|
trafficking protein particle complex 11 |
| chr1_+_76251879 | 0.27 |
ENST00000535300.1
ENST00000319942.3 |
RABGGTB
|
Rab geranylgeranyltransferase, beta subunit |
| chr10_+_70715884 | 0.27 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr2_-_175351744 | 0.27 |
ENST00000295500.4
ENST00000392552.2 ENST00000392551.2 |
GPR155
|
G protein-coupled receptor 155 |
| chr1_-_113498943 | 0.27 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr6_-_41040268 | 0.27 |
ENST00000373154.2
ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr17_-_18585541 | 0.27 |
ENST00000285274.5
ENST00000545289.1 ENST00000580145.1 |
ZNF286B
|
zinc finger protein 286B |
| chr13_+_103451548 | 0.27 |
ENST00000419638.1
|
BIVM
|
basic, immunoglobulin-like variable motif containing |
| chr6_+_19837592 | 0.27 |
ENST00000378700.3
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr1_+_39547082 | 0.27 |
ENST00000602421.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chrY_+_2803322 | 0.27 |
ENST00000383052.1
ENST00000155093.3 ENST00000449237.1 ENST00000443793.1 |
ZFY
|
zinc finger protein, Y-linked |
| chr6_+_41040678 | 0.27 |
ENST00000341376.6
ENST00000353205.5 |
NFYA
|
nuclear transcription factor Y, alpha |
| chr17_-_71258019 | 0.27 |
ENST00000344935.4
|
CPSF4L
|
cleavage and polyadenylation specific factor 4-like |
| chr12_+_104697504 | 0.26 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr14_+_35761580 | 0.26 |
ENST00000553809.1
ENST00000555764.1 ENST00000556506.1 |
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6 |
| chr11_-_46142615 | 0.26 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr3_+_133293278 | 0.26 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr6_-_74233480 | 0.26 |
ENST00000455918.1
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr15_-_52263937 | 0.26 |
ENST00000315141.5
ENST00000299601.5 |
LEO1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
| chr11_-_8986474 | 0.26 |
ENST00000525069.1
|
TMEM9B
|
TMEM9 domain family, member B |
Network of associatons between targets according to the STRING database.
First level regulatory network of RARA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 1.0 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.2 | 0.7 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.2 | 0.6 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.6 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 0.5 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.7 | GO:0006311 | meiotic gene conversion(GO:0006311) regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.6 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.1 | 0.4 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.1 | 0.5 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.3 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.7 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 | 0.4 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.3 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.6 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.1 | 0.3 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.1 | 0.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.3 | GO:0097460 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.2 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) |
| 0.1 | 0.6 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.4 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
| 0.1 | 0.4 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.8 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.5 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.4 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.1 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.7 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.1 | 1.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.3 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.4 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.1 | 0.6 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.3 | GO:1901073 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.5 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.1 | 0.2 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 0.5 | GO:0060702 | negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) |
| 0.1 | 1.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.2 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 0.1 | 0.5 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.2 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.6 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.7 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.4 | GO:0031620 | smooth muscle adaptation(GO:0014805) regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 1.3 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) positive regulation of penile erection(GO:0060406) |
| 0.0 | 1.1 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.3 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.2 | GO:0051836 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:2000410 | regulation of thymocyte migration(GO:2000410) |
| 0.0 | 1.0 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.4 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.3 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.5 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 1.1 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.2 | GO:0035815 | positive regulation of renal sodium excretion(GO:0035815) |
| 0.0 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.4 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.7 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.2 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.3 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.0 | 0.2 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.4 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.4 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.5 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.4 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.7 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0060809 | cardiac right atrium morphogenesis(GO:0003213) chemoattraction of serotonergic neuron axon(GO:0036517) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) chemoattraction of axon(GO:0061642) melanocyte proliferation(GO:0097325) negative regulation of cell proliferation in midbrain(GO:1904934) planar cell polarity pathway involved in midbrain dopaminergic neuron differentiation(GO:1904955) |
| 0.0 | 0.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.7 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 1.0 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.9 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.1 | GO:0001808 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.2 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.3 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.0 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.8 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.4 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.4 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.2 | 1.0 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 1.0 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.2 | 1.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 0.5 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 1.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 1.4 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.1 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.7 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 1.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.6 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 1.0 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.2 | 0.7 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.2 | 0.7 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.2 | 1.0 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.2 | 1.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 0.5 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) leukemia inhibitory factor receptor activity(GO:0004923) interleukin-6 binding(GO:0019981) |
| 0.1 | 0.7 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.3 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.6 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.3 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.5 | GO:0017089 | glycolipid transporter activity(GO:0017089) ceramide binding(GO:0097001) |
| 0.1 | 0.6 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.4 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 0.3 | GO:0005365 | myo-inositol transmembrane transporter activity(GO:0005365) |
| 0.1 | 0.3 | GO:0047661 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.2 | GO:0048244 | phytanoyl-CoA dioxygenase activity(GO:0048244) |
| 0.1 | 0.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.8 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.2 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.1 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.5 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.1 | 0.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.4 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.7 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 1.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0035529 | phosphodiesterase I activity(GO:0004528) NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.4 | GO:0016896 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.3 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.7 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0005115 | receptor tyrosine kinase-like orphan receptor binding(GO:0005115) chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.8 | GO:0019840 | retinoid binding(GO:0005501) isoprenoid binding(GO:0019840) |
| 0.0 | 0.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.9 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 1.2 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.0 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 1.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.3 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.0 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |