Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
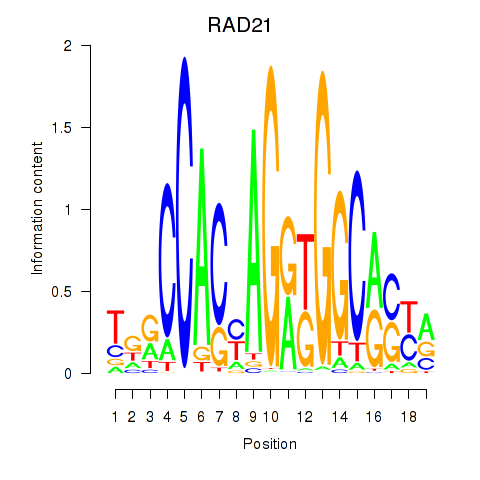
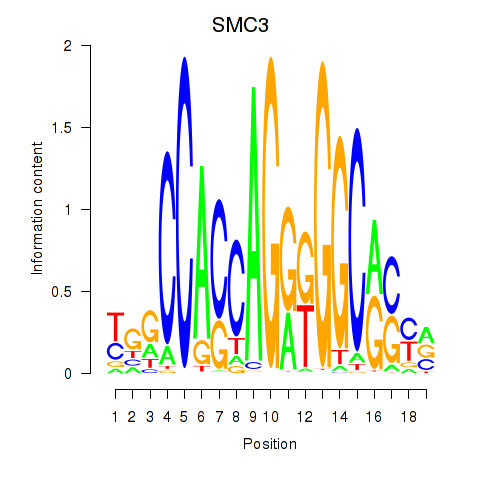
Results for RAD21_SMC3
Z-value: 1.08
Transcription factors associated with RAD21_SMC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RAD21
|
ENSG00000164754.8 | RAD21 cohesin complex component |
|
SMC3
|
ENSG00000108055.9 | structural maintenance of chromosomes 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RAD21 | hg19_v2_chr8_-_117886563_117886605 | -0.63 | 1.8e-01 | Click! |
| SMC3 | hg19_v2_chr10_+_112327425_112327516 | -0.56 | 2.5e-01 | Click! |
Activity profile of RAD21_SMC3 motif
Sorted Z-values of RAD21_SMC3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_109642799 | 1.13 |
ENST00000602755.1
|
SCARNA2
|
small Cajal body-specific RNA 2 |
| chr9_-_134615326 | 1.00 |
ENST00000438647.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr2_+_11273179 | 0.94 |
ENST00000381585.3
ENST00000405022.3 |
C2orf50
|
chromosome 2 open reading frame 50 |
| chrX_-_27417088 | 0.86 |
ENST00000608735.1
ENST00000422048.1 |
RP11-268G12.1
|
RP11-268G12.1 |
| chr11_+_73675873 | 0.85 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr3_+_50192457 | 0.80 |
ENST00000414301.1
ENST00000450338.1 |
SEMA3F
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr4_-_175041663 | 0.79 |
ENST00000503140.1
|
RP11-148L24.1
|
RP11-148L24.1 |
| chr6_+_31105426 | 0.76 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr14_-_75079294 | 0.74 |
ENST00000556359.1
|
LTBP2
|
latent transforming growth factor beta binding protein 2 |
| chr7_-_27219632 | 0.72 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr2_+_232316906 | 0.72 |
ENST00000370380.2
|
AC017104.2
|
Uncharacterized protein |
| chr18_+_43753500 | 0.72 |
ENST00000587591.1
ENST00000588730.1 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr12_-_51663682 | 0.72 |
ENST00000603838.1
|
SMAGP
|
small cell adhesion glycoprotein |
| chr8_-_11325047 | 0.70 |
ENST00000531804.1
|
FAM167A
|
family with sequence similarity 167, member A |
| chr19_+_10527449 | 0.67 |
ENST00000592685.1
ENST00000380702.2 |
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr1_+_840205 | 0.67 |
ENST00000607769.1
|
RP11-54O7.16
|
RP11-54O7.16 |
| chr17_-_26220366 | 0.66 |
ENST00000460380.2
ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9
RP1-66C13.4
|
LYR motif containing 9 Uncharacterized protein |
| chr6_-_41747595 | 0.66 |
ENST00000373018.3
|
FRS3
|
fibroblast growth factor receptor substrate 3 |
| chr7_-_100171270 | 0.65 |
ENST00000538735.1
|
SAP25
|
Sin3A-associated protein, 25kDa |
| chr8_-_146012660 | 0.59 |
ENST00000343459.4
ENST00000429371.2 ENST00000534445.1 |
ZNF34
|
zinc finger protein 34 |
| chr2_+_20866424 | 0.57 |
ENST00000272224.3
|
GDF7
|
growth differentiation factor 7 |
| chr12_+_49208234 | 0.57 |
ENST00000540990.1
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr9_+_116917807 | 0.56 |
ENST00000356083.3
|
COL27A1
|
collagen, type XXVII, alpha 1 |
| chr10_-_76995675 | 0.54 |
ENST00000469299.1
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr19_+_4791722 | 0.54 |
ENST00000269856.3
|
FEM1A
|
fem-1 homolog a (C. elegans) |
| chr1_-_156675535 | 0.54 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr1_-_145589424 | 0.54 |
ENST00000334513.5
|
NUDT17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr9_+_33617760 | 0.52 |
ENST00000379435.3
|
TRBV20OR9-2
|
T cell receptor beta variable 20/OR9-2 (non-functional) |
| chr11_-_130786333 | 0.50 |
ENST00000533214.1
ENST00000528555.1 ENST00000530356.1 ENST00000539184.1 |
SNX19
|
sorting nexin 19 |
| chr11_+_2421718 | 0.50 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr7_-_27219849 | 0.50 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr17_-_37308824 | 0.49 |
ENST00000415163.1
ENST00000441877.1 ENST00000444911.2 |
PLXDC1
|
plexin domain containing 1 |
| chr12_+_110338323 | 0.48 |
ENST00000312777.5
ENST00000536408.2 |
TCHP
|
trichoplein, keratin filament binding |
| chr16_+_69345243 | 0.47 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr20_+_36531544 | 0.47 |
ENST00000448944.1
|
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr8_-_27850141 | 0.46 |
ENST00000524352.1
|
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr1_-_156675564 | 0.46 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr11_-_85565906 | 0.45 |
ENST00000544076.1
|
AP000974.1
|
CDNA FLJ26432 fis, clone KDN01418; Uncharacterized protein |
| chr8_-_11873043 | 0.44 |
ENST00000527396.1
|
RP11-481A20.11
|
Protein LOC101060662 |
| chrX_-_131353461 | 0.44 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr6_-_18122843 | 0.44 |
ENST00000340650.3
|
NHLRC1
|
NHL repeat containing E3 ubiquitin protein ligase 1 |
| chr11_+_58938903 | 0.44 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr18_-_26941900 | 0.44 |
ENST00000577674.1
|
CTD-2515C13.2
|
CTD-2515C13.2 |
| chr5_+_133859996 | 0.43 |
ENST00000512386.1
|
PHF15
|
jade family PHD finger 2 |
| chr19_-_55653259 | 0.43 |
ENST00000593194.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr16_-_19897455 | 0.43 |
ENST00000568214.1
ENST00000569479.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr17_-_79827808 | 0.43 |
ENST00000580685.1
|
ARHGDIA
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr19_-_49339080 | 0.42 |
ENST00000595764.1
|
HSD17B14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr19_-_1848451 | 0.41 |
ENST00000170168.4
|
REXO1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae) |
| chr16_-_15950868 | 0.41 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr12_+_110338063 | 0.41 |
ENST00000405876.4
|
TCHP
|
trichoplein, keratin filament binding |
| chr1_+_154966058 | 0.40 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr20_-_3762087 | 0.40 |
ENST00000379756.3
|
SPEF1
|
sperm flagellar 1 |
| chr19_-_6481776 | 0.40 |
ENST00000543576.1
ENST00000590173.1 ENST00000381480.2 |
DENND1C
|
DENN/MADD domain containing 1C |
| chr12_+_58176525 | 0.40 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr6_-_139309378 | 0.39 |
ENST00000450536.2
ENST00000258062.5 |
REPS1
|
RALBP1 associated Eps domain containing 1 |
| chr11_+_46368956 | 0.38 |
ENST00000543978.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr19_-_4400415 | 0.38 |
ENST00000598564.1
ENST00000417295.2 ENST00000269886.3 |
SH3GL1
|
SH3-domain GRB2-like 1 |
| chr14_+_23305783 | 0.38 |
ENST00000547279.1
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr22_-_19132154 | 0.38 |
ENST00000252137.6
|
DGCR14
|
DiGeorge syndrome critical region gene 14 |
| chr11_-_65359947 | 0.38 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chr1_-_6761855 | 0.38 |
ENST00000426784.1
ENST00000377573.5 ENST00000377577.5 ENST00000294401.7 |
DNAJC11
|
DnaJ (Hsp40) homolog, subfamily C, member 11 |
| chr1_-_159915386 | 0.38 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr8_+_28480246 | 0.37 |
ENST00000523149.1
|
EXTL3
|
exostosin-like glycosyltransferase 3 |
| chr11_+_120039685 | 0.37 |
ENST00000530303.1
ENST00000319763.1 |
AP000679.2
|
Uncharacterized protein |
| chr5_-_33297946 | 0.37 |
ENST00000510327.1
|
CTD-2066L21.3
|
CTD-2066L21.3 |
| chr20_-_34542548 | 0.37 |
ENST00000305978.2
|
SCAND1
|
SCAN domain containing 1 |
| chr5_-_140998616 | 0.37 |
ENST00000389054.3
ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1
|
diaphanous-related formin 1 |
| chr20_+_43992094 | 0.37 |
ENST00000453003.1
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr1_+_20208870 | 0.37 |
ENST00000375120.3
|
OTUD3
|
OTU domain containing 3 |
| chr19_-_58400148 | 0.36 |
ENST00000595048.1
ENST00000600634.1 ENST00000595295.1 ENST00000596604.1 ENST00000597342.1 ENST00000597807.1 |
ZNF814
|
zinc finger protein 814 |
| chr1_-_35325400 | 0.36 |
ENST00000521580.2
|
SMIM12
|
small integral membrane protein 12 |
| chr20_-_30311703 | 0.35 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr10_-_76995769 | 0.34 |
ENST00000372538.3
|
COMTD1
|
catechol-O-methyltransferase domain containing 1 |
| chr8_-_48651648 | 0.34 |
ENST00000408965.3
|
CEBPD
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr17_-_73511504 | 0.34 |
ENST00000581870.1
|
CASKIN2
|
CASK interacting protein 2 |
| chr19_+_56165480 | 0.34 |
ENST00000450554.2
|
U2AF2
|
U2 small nuclear RNA auxiliary factor 2 |
| chr5_-_140998481 | 0.34 |
ENST00000518047.1
|
DIAPH1
|
diaphanous-related formin 1 |
| chr2_-_74710078 | 0.33 |
ENST00000290418.4
|
CCDC142
|
coiled-coil domain containing 142 |
| chrX_+_49969405 | 0.33 |
ENST00000376042.1
|
CCNB3
|
cyclin B3 |
| chr16_+_30759844 | 0.33 |
ENST00000565897.1
|
PHKG2
|
phosphorylase kinase, gamma 2 (testis) |
| chr19_-_4791219 | 0.33 |
ENST00000598782.1
|
AC005523.3
|
AC005523.3 |
| chr4_-_41884620 | 0.33 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr19_-_3547305 | 0.33 |
ENST00000589063.1
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr14_-_93582148 | 0.33 |
ENST00000267615.6
ENST00000553452.1 |
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr19_+_49496705 | 0.33 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr16_-_2379688 | 0.32 |
ENST00000567910.1
|
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr14_+_55738021 | 0.32 |
ENST00000313833.4
|
FBXO34
|
F-box protein 34 |
| chr15_+_75074410 | 0.32 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr1_+_151129135 | 0.32 |
ENST00000602841.1
|
SCNM1
|
sodium channel modifier 1 |
| chr12_-_95510743 | 0.32 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr10_+_104154229 | 0.32 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr16_+_90086002 | 0.32 |
ENST00000563936.1
ENST00000536122.1 ENST00000561675.1 |
GAS8
|
growth arrest-specific 8 |
| chr19_+_36027660 | 0.32 |
ENST00000585510.1
|
GAPDHS
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr19_+_5823813 | 0.32 |
ENST00000303212.2
|
NRTN
|
neurturin |
| chr11_-_130786400 | 0.32 |
ENST00000265909.4
|
SNX19
|
sorting nexin 19 |
| chr9_-_123239632 | 0.31 |
ENST00000416449.1
|
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr10_-_79789291 | 0.31 |
ENST00000372371.3
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr14_+_23305760 | 0.31 |
ENST00000311852.6
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted) |
| chr2_+_239067597 | 0.31 |
ENST00000546354.1
|
FAM132B
|
family with sequence similarity 132, member B |
| chrX_+_46772065 | 0.31 |
ENST00000455411.1
|
PHF16
|
jade family PHD finger 3 |
| chr17_+_41561317 | 0.31 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr11_+_46369077 | 0.31 |
ENST00000456247.2
ENST00000421244.2 ENST00000318201.8 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr16_-_2390704 | 0.31 |
ENST00000301732.5
ENST00000382381.3 |
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr12_-_7261772 | 0.30 |
ENST00000545280.1
ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL
|
complement component 1, r subcomponent-like |
| chr14_+_105190514 | 0.30 |
ENST00000330877.2
|
ADSSL1
|
adenylosuccinate synthase like 1 |
| chr12_+_50898881 | 0.30 |
ENST00000301180.5
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr20_-_1447547 | 0.30 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr9_-_139940608 | 0.30 |
ENST00000371601.4
|
NPDC1
|
neural proliferation, differentiation and control, 1 |
| chr22_+_22901750 | 0.30 |
ENST00000407120.1
|
LL22NC03-63E9.3
|
Uncharacterized protein |
| chr20_+_48807351 | 0.30 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr20_+_36531499 | 0.30 |
ENST00000373458.3
ENST00000373461.4 ENST00000373459.4 |
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr19_-_49496557 | 0.29 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr17_-_73901494 | 0.29 |
ENST00000309352.3
|
MRPL38
|
mitochondrial ribosomal protein L38 |
| chr4_+_7045042 | 0.29 |
ENST00000310074.7
ENST00000512388.1 |
TADA2B
|
transcriptional adaptor 2B |
| chr19_+_49496782 | 0.29 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr19_+_47852538 | 0.28 |
ENST00000328771.4
|
DHX34
|
DEAH (Asp-Glu-Ala-His) box polypeptide 34 |
| chr1_-_186344802 | 0.28 |
ENST00000451586.1
|
TPR
|
translocated promoter region, nuclear basket protein |
| chr9_-_139258235 | 0.28 |
ENST00000371738.3
|
DNLZ
|
DNL-type zinc finger |
| chr17_-_43209862 | 0.28 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr11_-_66104237 | 0.28 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr9_+_103189536 | 0.28 |
ENST00000374885.1
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr1_-_146644036 | 0.28 |
ENST00000425272.2
|
PRKAB2
|
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
| chr12_-_1058685 | 0.28 |
ENST00000397230.2
ENST00000542785.1 ENST00000544742.1 ENST00000536177.1 ENST00000539046.1 ENST00000541619.1 |
RAD52
|
RAD52 homolog (S. cerevisiae) |
| chr19_+_7445850 | 0.28 |
ENST00000593531.1
|
CTD-2207O23.3
|
Rho guanine nucleotide exchange factor 18 |
| chr2_+_166095898 | 0.27 |
ENST00000424833.1
ENST00000375437.2 ENST00000357398.3 |
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit |
| chr11_+_61447845 | 0.27 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr14_-_93582214 | 0.27 |
ENST00000556603.2
ENST00000354313.3 |
ITPK1
|
inositol-tetrakisphosphate 1-kinase |
| chr1_-_139379 | 0.27 |
ENST00000423372.3
|
AL627309.1
|
Uncharacterized protein |
| chr5_+_137688285 | 0.27 |
ENST00000314358.5
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr1_+_146714291 | 0.27 |
ENST00000431239.1
ENST00000369259.3 ENST00000369258.4 ENST00000361293.5 |
CHD1L
|
chromodomain helicase DNA binding protein 1-like |
| chr17_-_73511584 | 0.27 |
ENST00000321617.3
|
CASKIN2
|
CASK interacting protein 2 |
| chr16_+_15737124 | 0.27 |
ENST00000396355.1
ENST00000396353.2 |
NDE1
|
nudE neurodevelopment protein 1 |
| chr16_+_66638685 | 0.27 |
ENST00000565003.1
|
CMTM3
|
CKLF-like MARVEL transmembrane domain containing 3 |
| chr11_+_46368975 | 0.27 |
ENST00000527911.1
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr16_+_30406721 | 0.27 |
ENST00000320159.2
|
ZNF48
|
zinc finger protein 48 |
| chr10_-_15413035 | 0.27 |
ENST00000378116.4
ENST00000455654.1 |
FAM171A1
|
family with sequence similarity 171, member A1 |
| chr11_-_17410629 | 0.27 |
ENST00000526912.1
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11 |
| chr1_+_36348790 | 0.27 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr1_+_178511876 | 0.27 |
ENST00000367638.1
ENST00000367636.4 |
C1orf220
C1ORF220
|
chromosome 1 open reading frame 220 C1ORF220 |
| chr20_+_3801162 | 0.27 |
ENST00000379573.2
ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr9_-_134615443 | 0.27 |
ENST00000372195.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr16_+_2255447 | 0.27 |
ENST00000562352.1
ENST00000562479.1 ENST00000301724.10 |
MLST8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr5_+_172571445 | 0.27 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr16_-_790982 | 0.26 |
ENST00000301694.5
ENST00000251588.2 |
NARFL
|
nuclear prelamin A recognition factor-like |
| chr1_+_10003486 | 0.26 |
ENST00000403197.1
ENST00000377205.1 |
NMNAT1
|
nicotinamide nucleotide adenylyltransferase 1 |
| chr11_-_8285405 | 0.26 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr10_-_71906342 | 0.26 |
ENST00000287078.6
ENST00000335494.5 |
TYSND1
|
trypsin domain containing 1 |
| chr8_-_145550571 | 0.26 |
ENST00000332324.4
|
DGAT1
|
diacylglycerol O-acyltransferase 1 |
| chr16_+_1756162 | 0.26 |
ENST00000250894.4
ENST00000356010.5 |
MAPK8IP3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr6_-_2634820 | 0.26 |
ENST00000296847.3
|
C6orf195
|
chromosome 6 open reading frame 195 |
| chr12_-_56583332 | 0.25 |
ENST00000347471.4
ENST00000267064.4 ENST00000394023.3 |
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr19_-_58400372 | 0.25 |
ENST00000597832.1
ENST00000435989.2 |
ZNF814
|
zinc finger protein 814 |
| chr19_+_18043810 | 0.25 |
ENST00000445755.2
|
CCDC124
|
coiled-coil domain containing 124 |
| chr17_-_1090599 | 0.25 |
ENST00000544583.2
|
ABR
|
active BCR-related |
| chr7_-_5569588 | 0.25 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr7_-_99679324 | 0.25 |
ENST00000292393.5
ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3
|
zinc finger protein 3 |
| chr12_-_56583243 | 0.25 |
ENST00000550164.1
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr15_+_75074385 | 0.25 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr15_-_64338521 | 0.25 |
ENST00000457488.1
ENST00000558069.1 |
DAPK2
|
death-associated protein kinase 2 |
| chr14_-_102701740 | 0.25 |
ENST00000561150.1
ENST00000522867.1 |
MOK
|
MOK protein kinase |
| chr5_-_125930877 | 0.25 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr8_-_27850180 | 0.25 |
ENST00000380385.2
ENST00000301906.4 ENST00000354914.3 |
SCARA5
|
scavenger receptor class A, member 5 (putative) |
| chr10_+_81107216 | 0.24 |
ENST00000394579.3
ENST00000225174.3 |
PPIF
|
peptidylprolyl isomerase F |
| chr12_+_7033616 | 0.24 |
ENST00000356654.4
|
ATN1
|
atrophin 1 |
| chr12_-_15374343 | 0.24 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr1_-_212208842 | 0.24 |
ENST00000366992.3
ENST00000366993.3 ENST00000440600.2 ENST00000366994.3 |
INTS7
|
integrator complex subunit 7 |
| chr2_+_234263120 | 0.24 |
ENST00000264057.2
ENST00000427930.1 |
DGKD
|
diacylglycerol kinase, delta 130kDa |
| chr17_-_34890732 | 0.24 |
ENST00000268852.9
|
MYO19
|
myosin XIX |
| chr17_-_33446735 | 0.24 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr12_+_51985001 | 0.24 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr9_+_133884469 | 0.24 |
ENST00000361069.4
|
LAMC3
|
laminin, gamma 3 |
| chr17_-_34890759 | 0.24 |
ENST00000431794.3
|
MYO19
|
myosin XIX |
| chr9_-_38069208 | 0.23 |
ENST00000377707.3
ENST00000377700.4 |
SHB
|
Src homology 2 domain containing adaptor protein B |
| chr5_-_125930929 | 0.23 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr16_-_23568651 | 0.23 |
ENST00000563232.1
ENST00000563459.1 ENST00000449606.1 |
EARS2
|
glutamyl-tRNA synthetase 2, mitochondrial |
| chr1_-_40367668 | 0.23 |
ENST00000397332.2
ENST00000429311.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr5_-_175461683 | 0.23 |
ENST00000514861.1
|
THOC3
|
THO complex 3 |
| chr7_-_4923259 | 0.23 |
ENST00000536091.1
|
RADIL
|
Ras association and DIL domains |
| chr6_+_33359582 | 0.23 |
ENST00000450504.1
|
KIFC1
|
kinesin family member C1 |
| chr12_-_82752159 | 0.23 |
ENST00000552377.1
|
CCDC59
|
coiled-coil domain containing 59 |
| chr7_-_91509972 | 0.23 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr16_+_22308717 | 0.23 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr11_-_68611721 | 0.23 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr6_+_138188378 | 0.23 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr19_-_55652290 | 0.23 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr20_-_1447467 | 0.23 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr9_+_130965677 | 0.22 |
ENST00000393594.3
ENST00000486160.1 |
DNM1
|
dynamin 1 |
| chr1_+_154244987 | 0.22 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr8_-_145911188 | 0.22 |
ENST00000540274.1
|
ARHGAP39
|
Rho GTPase activating protein 39 |
| chr22_-_22292934 | 0.22 |
ENST00000538191.1
ENST00000424647.1 ENST00000407142.1 |
PPM1F
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chrX_+_153029633 | 0.22 |
ENST00000538966.1
ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3
|
plexin B3 |
| chr15_-_43882353 | 0.22 |
ENST00000453080.1
ENST00000360301.4 ENST00000360135.4 ENST00000417085.1 ENST00000431962.1 ENST00000334933.4 ENST00000381879.4 ENST00000420765.1 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr11_+_8008867 | 0.22 |
ENST00000309828.4
ENST00000449102.2 |
EIF3F
|
eukaryotic translation initiation factor 3, subunit F |
| chr20_+_57427765 | 0.21 |
ENST00000371100.4
|
GNAS
|
GNAS complex locus |
| chr7_-_4923315 | 0.21 |
ENST00000399583.3
|
RADIL
|
Ras association and DIL domains |
| chr5_+_176514413 | 0.21 |
ENST00000513166.1
|
FGFR4
|
fibroblast growth factor receptor 4 |
| chr6_+_155054459 | 0.21 |
ENST00000367178.3
ENST00000417268.1 ENST00000367186.4 |
SCAF8
|
SR-related CTD-associated factor 8 |
| chr16_-_30997533 | 0.21 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr19_+_12949251 | 0.21 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of RAD21_SMC3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.2 | 0.9 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 0.5 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 0.6 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.4 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.5 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.3 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.3 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.6 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.3 | GO:2000349 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.3 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.1 | 0.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.4 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.1 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.6 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.2 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.3 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.6 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) |
| 0.1 | 0.2 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.1 | 0.5 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 | 0.7 | GO:1990834 | response to odorant(GO:1990834) |
| 0.1 | 0.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.2 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.4 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 1.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.9 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 1.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.2 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 1.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.2 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.5 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0033242 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.7 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.2 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.3 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.3 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.9 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.0 | 0.1 | GO:0035981 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.0 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0075528 | negative regulation of extracellular matrix disassembly(GO:0010716) induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.0 | 0.3 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.3 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.1 | GO:0002586 | regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.4 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.2 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.5 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 1.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.3 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.4 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.2 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.9 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:1904387 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.5 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.6 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.4 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.3 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.4 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.0 | 0.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.7 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.0 | GO:0060268 | respiratory burst involved in inflammatory response(GO:0002536) regulation of respiratory burst involved in inflammatory response(GO:0060264) positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.0 | GO:2000627 | regulation of miRNA catabolic process(GO:2000625) positive regulation of miRNA catabolic process(GO:2000627) |
| 0.0 | 0.1 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.3 | GO:0051197 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.6 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.3 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.1 | GO:1902074 | response to salt(GO:1902074) |
| 0.0 | 0.0 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.2 | GO:0060537 | muscle tissue development(GO:0060537) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.5 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.5 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.0 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0032059 | muscle thin filament tropomyosin(GO:0005862) bleb(GO:0032059) |
| 0.0 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.0 | GO:0097124 | Y chromosome(GO:0000806) cyclin A1-CDK2 complex(GO:0097123) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 1.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0052831 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.2 | 0.5 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 1.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.4 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 0.3 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.1 | 0.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.4 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 1.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.4 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.1 | 0.2 | GO:0048257 | 3'-flap endonuclease activity(GO:0048257) |
| 0.1 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 1.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 1.0 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.0 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.0 | 0.9 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.2 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 1.1 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.1 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.6 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |