Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for PTF1A
Z-value: 0.69
Transcription factors associated with PTF1A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PTF1A
|
ENSG00000168267.5 | pancreas associated transcription factor 1a |
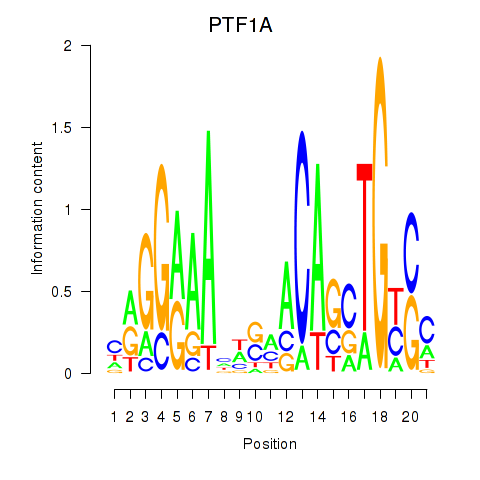
Activity profile of PTF1A motif
Sorted Z-values of PTF1A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_2912363 | 1.30 |
ENST00000544366.1
|
FKBP4
|
FK506 binding protein 4, 59kDa |
| chr11_-_1606513 | 0.71 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chr15_+_81225699 | 0.68 |
ENST00000560027.1
|
KIAA1199
|
KIAA1199 |
| chr12_-_57644952 | 0.55 |
ENST00000554578.1
ENST00000546246.2 ENST00000553489.1 ENST00000332782.2 |
STAC3
|
SH3 and cysteine rich domain 3 |
| chr17_+_68047418 | 0.55 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr3_-_52868931 | 0.50 |
ENST00000486659.1
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr3_-_52869205 | 0.43 |
ENST00000446157.2
|
MUSTN1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr19_+_29493456 | 0.43 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr8_-_145642203 | 0.40 |
ENST00000526658.1
|
SLC39A4
|
solute carrier family 39 (zinc transporter), member 4 |
| chr1_-_45956868 | 0.38 |
ENST00000451835.2
|
TESK2
|
testis-specific kinase 2 |
| chr13_-_111522162 | 0.38 |
ENST00000538077.1
|
LINC00346
|
long intergenic non-protein coding RNA 346 |
| chr16_-_75569068 | 0.36 |
ENST00000336257.3
ENST00000565039.1 |
CHST5
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 |
| chr19_+_6464502 | 0.36 |
ENST00000308243.7
|
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr5_+_56205878 | 0.35 |
ENST00000423328.1
|
SETD9
|
SET domain containing 9 |
| chr15_+_84841242 | 0.33 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr16_+_89696692 | 0.32 |
ENST00000261615.4
|
DPEP1
|
dipeptidase 1 (renal) |
| chr19_+_11670245 | 0.29 |
ENST00000585493.1
|
ZNF627
|
zinc finger protein 627 |
| chr4_-_175041663 | 0.26 |
ENST00000503140.1
|
RP11-148L24.1
|
RP11-148L24.1 |
| chr1_-_114447620 | 0.25 |
ENST00000369569.1
ENST00000432415.1 ENST00000369571.2 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr19_+_6464243 | 0.24 |
ENST00000600229.1
ENST00000356762.3 |
CRB3
|
crumbs homolog 3 (Drosophila) |
| chr5_-_55902050 | 0.24 |
ENST00000438651.1
|
AC022431.2
|
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
| chr7_-_99097863 | 0.24 |
ENST00000426306.2
ENST00000337673.6 |
ZNF394
|
zinc finger protein 394 |
| chr1_-_78149041 | 0.24 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr19_-_48389651 | 0.22 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr18_-_5238525 | 0.22 |
ENST00000581170.1
ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5
LINC00526
|
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr3_-_155011483 | 0.21 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr1_-_45956822 | 0.21 |
ENST00000372086.3
ENST00000341771.6 |
TESK2
|
testis-specific kinase 2 |
| chr8_+_97773202 | 0.20 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr3_+_167453026 | 0.20 |
ENST00000472941.1
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr16_-_2004683 | 0.20 |
ENST00000268661.7
|
RPL3L
|
ribosomal protein L3-like |
| chr1_-_9129735 | 0.19 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr8_-_22089845 | 0.19 |
ENST00000454243.2
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr3_-_172241250 | 0.18 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr2_-_95831158 | 0.18 |
ENST00000447814.1
|
ZNF514
|
zinc finger protein 514 |
| chrX_-_13956737 | 0.18 |
ENST00000454189.2
|
GPM6B
|
glycoprotein M6B |
| chr3_-_69249863 | 0.17 |
ENST00000478263.1
ENST00000462512.1 |
FRMD4B
|
FERM domain containing 4B |
| chr1_+_114447763 | 0.17 |
ENST00000369563.3
|
DCLRE1B
|
DNA cross-link repair 1B |
| chr8_-_135725205 | 0.17 |
ENST00000523399.1
ENST00000377838.3 |
ZFAT
|
zinc finger and AT hook domain containing |
| chrX_-_21676442 | 0.17 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr1_-_114447412 | 0.17 |
ENST00000369567.1
ENST00000369566.3 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr8_+_97773457 | 0.16 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr17_-_15175604 | 0.16 |
ENST00000431343.1
|
AC005703.3
|
AC005703.3 |
| chr3_+_49027308 | 0.16 |
ENST00000383729.4
ENST00000343546.4 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr1_-_9129631 | 0.15 |
ENST00000377414.3
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr8_-_22089533 | 0.15 |
ENST00000321613.3
|
PHYHIP
|
phytanoyl-CoA 2-hydroxylase interacting protein |
| chr10_-_43633747 | 0.15 |
ENST00000609407.1
|
RP11-351D16.3
|
RP11-351D16.3 |
| chr8_+_11291429 | 0.13 |
ENST00000533578.1
|
C8orf12
|
chromosome 8 open reading frame 12 |
| chr5_-_16916624 | 0.13 |
ENST00000513882.1
|
MYO10
|
myosin X |
| chr20_+_42544782 | 0.13 |
ENST00000423191.2
ENST00000372999.1 |
TOX2
|
TOX high mobility group box family member 2 |
| chr6_-_56258892 | 0.13 |
ENST00000370819.1
|
COL21A1
|
collagen, type XXI, alpha 1 |
| chr19_+_17326521 | 0.13 |
ENST00000593597.1
|
USE1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr9_-_97090926 | 0.13 |
ENST00000335456.7
ENST00000253262.4 ENST00000341207.4 |
NUTM2F
|
NUT family member 2F |
| chr4_+_184826418 | 0.12 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr12_+_12966250 | 0.12 |
ENST00000352940.4
ENST00000358007.3 ENST00000544400.1 |
DDX47
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
| chr17_+_40996590 | 0.12 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr10_-_28571015 | 0.12 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr1_-_114447683 | 0.12 |
ENST00000256658.4
ENST00000369564.1 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr19_+_54495542 | 0.12 |
ENST00000252729.2
ENST00000352529.1 |
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr8_-_38126675 | 0.11 |
ENST00000531823.1
ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr17_+_67498538 | 0.11 |
ENST00000589647.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr11_-_111794446 | 0.11 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr13_+_49794474 | 0.11 |
ENST00000218721.1
ENST00000398307.1 |
MLNR
|
motilin receptor |
| chr18_+_5238055 | 0.11 |
ENST00000582363.1
ENST00000582008.1 ENST00000580082.1 |
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr1_-_45956800 | 0.11 |
ENST00000538496.1
|
TESK2
|
testis-specific kinase 2 |
| chr12_-_50677255 | 0.11 |
ENST00000551691.1
ENST00000394943.3 ENST00000341247.4 |
LIMA1
|
LIM domain and actin binding 1 |
| chr19_-_46526304 | 0.11 |
ENST00000008938.4
|
PGLYRP1
|
peptidoglycan recognition protein 1 |
| chr2_+_183943464 | 0.11 |
ENST00000354221.4
|
DUSP19
|
dual specificity phosphatase 19 |
| chr4_-_14889791 | 0.11 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr8_+_32405728 | 0.11 |
ENST00000523079.1
ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1
|
neuregulin 1 |
| chr16_-_15950868 | 0.10 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr3_+_49027771 | 0.10 |
ENST00000475629.1
ENST00000444213.1 |
P4HTM
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr11_-_45687128 | 0.10 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr18_+_33877654 | 0.09 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr11_-_63933504 | 0.09 |
ENST00000255681.6
|
MACROD1
|
MACRO domain containing 1 |
| chr16_-_85146040 | 0.09 |
ENST00000539556.1
|
FAM92B
|
family with sequence similarity 92, member B |
| chr2_-_86422523 | 0.09 |
ENST00000442664.2
ENST00000409051.2 ENST00000449247.2 |
IMMT
|
inner membrane protein, mitochondrial |
| chr11_+_7597639 | 0.09 |
ENST00000533792.1
|
PPFIBP2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr13_+_113633620 | 0.09 |
ENST00000421756.1
ENST00000375601.3 |
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr20_+_18125727 | 0.09 |
ENST00000489634.2
|
CSRP2BP
|
CSRP2 binding protein |
| chr4_+_183370146 | 0.09 |
ENST00000510504.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr7_-_72993033 | 0.09 |
ENST00000305632.5
|
TBL2
|
transducin (beta)-like 2 |
| chr1_+_171454639 | 0.09 |
ENST00000392078.3
ENST00000426496.2 |
PRRC2C
|
proline-rich coiled-coil 2C |
| chr1_-_205649580 | 0.08 |
ENST00000367145.3
|
SLC45A3
|
solute carrier family 45, member 3 |
| chr11_-_130298888 | 0.08 |
ENST00000257359.6
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
| chr1_-_76076793 | 0.08 |
ENST00000370859.3
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr5_-_162887071 | 0.08 |
ENST00000302764.4
|
NUDCD2
|
NudC domain containing 2 |
| chrX_+_154299690 | 0.08 |
ENST00000340647.4
ENST00000330045.7 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr19_+_54496132 | 0.08 |
ENST00000346968.2
|
CACNG6
|
calcium channel, voltage-dependent, gamma subunit 6 |
| chr2_-_237172988 | 0.08 |
ENST00000409749.3
ENST00000430053.1 |
ASB18
|
ankyrin repeat and SOCS box containing 18 |
| chr8_+_32405785 | 0.07 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr19_-_10491130 | 0.07 |
ENST00000530829.1
ENST00000529370.1 |
TYK2
|
tyrosine kinase 2 |
| chr10_+_43633914 | 0.07 |
ENST00000374466.3
ENST00000374464.1 |
CSGALNACT2
|
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr5_+_140557371 | 0.07 |
ENST00000239444.2
|
PCDHB8
|
protocadherin beta 8 |
| chr17_-_42143963 | 0.07 |
ENST00000585388.1
ENST00000293406.3 |
LSM12
|
LSM12 homolog (S. cerevisiae) |
| chr4_+_40198527 | 0.07 |
ENST00000381799.5
|
RHOH
|
ras homolog family member H |
| chr9_+_4985228 | 0.07 |
ENST00000381652.3
|
JAK2
|
Janus kinase 2 |
| chr20_-_44937124 | 0.06 |
ENST00000537909.1
|
CDH22
|
cadherin 22, type 2 |
| chr2_+_202122703 | 0.06 |
ENST00000447616.1
ENST00000358485.4 |
CASP8
|
caspase 8, apoptosis-related cysteine peptidase |
| chrX_-_107019181 | 0.06 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr17_+_67498295 | 0.06 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr1_+_26856236 | 0.06 |
ENST00000374168.2
ENST00000374166.4 |
RPS6KA1
|
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr1_+_171454659 | 0.06 |
ENST00000367742.3
ENST00000338920.4 |
PRRC2C
|
proline-rich coiled-coil 2C |
| chr8_+_11351494 | 0.06 |
ENST00000259089.4
|
BLK
|
B lymphoid tyrosine kinase |
| chr12_-_90102594 | 0.06 |
ENST00000428670.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr12_-_3862347 | 0.06 |
ENST00000444507.1
|
EFCAB4B
|
EF-hand calcium binding domain 4B |
| chr8_-_38126635 | 0.06 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr11_+_64058758 | 0.06 |
ENST00000538767.1
|
KCNK4
|
potassium channel, subfamily K, member 4 |
| chr12_+_75784850 | 0.06 |
ENST00000550916.1
ENST00000435775.1 ENST00000378689.2 ENST00000378692.3 ENST00000320460.4 ENST00000547164.1 |
GLIPR1L2
|
GLI pathogenesis-related 1 like 2 |
| chr3_+_35721182 | 0.06 |
ENST00000413378.1
ENST00000417925.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr12_-_56223394 | 0.05 |
ENST00000317269.3
|
DNAJC14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr6_-_2962331 | 0.05 |
ENST00000380524.1
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr2_+_173600514 | 0.05 |
ENST00000264111.6
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_+_8704748 | 0.05 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr17_+_53342311 | 0.05 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr2_+_3393453 | 0.05 |
ENST00000441983.1
|
TRAPPC12
|
trafficking protein particle complex 12 |
| chr6_-_24877490 | 0.05 |
ENST00000540914.1
ENST00000378023.4 |
FAM65B
|
family with sequence similarity 65, member B |
| chr17_-_3461092 | 0.05 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr5_-_118324200 | 0.05 |
ENST00000515439.3
ENST00000510708.1 |
DTWD2
|
DTW domain containing 2 |
| chr13_+_107028897 | 0.05 |
ENST00000439790.1
ENST00000435024.1 |
LINC00460
|
long intergenic non-protein coding RNA 460 |
| chr2_+_173600565 | 0.05 |
ENST00000397081.3
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr1_+_40204538 | 0.05 |
ENST00000324379.5
ENST00000356511.2 ENST00000497370.1 ENST00000470213.1 ENST00000372835.5 ENST00000372830.1 |
PPIE
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr3_-_195938256 | 0.05 |
ENST00000296326.3
|
ZDHHC19
|
zinc finger, DHHC-type containing 19 |
| chr5_+_78532003 | 0.05 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr3_+_40518599 | 0.05 |
ENST00000314686.5
ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619
|
zinc finger protein 619 |
| chr16_+_2802316 | 0.04 |
ENST00000301740.8
|
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr17_-_61778528 | 0.04 |
ENST00000584645.1
|
LIMD2
|
LIM domain containing 2 |
| chr5_-_39425290 | 0.04 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr10_-_10504285 | 0.04 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr17_-_73892992 | 0.04 |
ENST00000540128.1
ENST00000269383.3 |
TRIM65
|
tripartite motif containing 65 |
| chr1_-_76076759 | 0.04 |
ENST00000370855.5
|
SLC44A5
|
solute carrier family 44, member 5 |
| chr6_-_24489842 | 0.04 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chrX_+_15808569 | 0.04 |
ENST00000380308.3
ENST00000307771.7 |
ZRSR2
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr7_-_152373216 | 0.04 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr11_-_61735103 | 0.04 |
ENST00000529191.1
ENST00000529631.1 ENST00000530019.1 ENST00000529548.1 ENST00000273550.7 |
FTH1
|
ferritin, heavy polypeptide 1 |
| chr6_-_127780510 | 0.04 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr16_+_2802623 | 0.03 |
ENST00000576924.1
ENST00000575009.1 ENST00000576415.1 ENST00000571378.1 |
SRRM2
|
serine/arginine repetitive matrix 2 |
| chr12_+_53689309 | 0.03 |
ENST00000351500.3
ENST00000550846.1 ENST00000334478.4 ENST00000549759.1 |
PFDN5
|
prefoldin subunit 5 |
| chr16_-_65155833 | 0.03 |
ENST00000566827.1
ENST00000394156.3 ENST00000562998.1 |
CDH11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr19_+_42300548 | 0.03 |
ENST00000344550.4
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr10_+_123970670 | 0.03 |
ENST00000496913.2
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr2_-_228497888 | 0.03 |
ENST00000264387.4
ENST00000409066.1 |
C2orf83
|
chromosome 2 open reading frame 83 |
| chr5_-_39425222 | 0.03 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr20_-_48732472 | 0.03 |
ENST00000340309.3
ENST00000415862.2 ENST00000371677.3 ENST00000420027.2 |
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr20_+_44441304 | 0.02 |
ENST00000352551.5
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr10_+_76871454 | 0.02 |
ENST00000372687.4
|
SAMD8
|
sterile alpha motif domain containing 8 |
| chr6_-_62996066 | 0.02 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr12_+_112451120 | 0.02 |
ENST00000261735.3
ENST00000455836.1 |
ERP29
|
endoplasmic reticulum protein 29 |
| chr12_+_50451331 | 0.02 |
ENST00000228468.4
|
ASIC1
|
acid-sensing (proton-gated) ion channel 1 |
| chr17_+_12692774 | 0.02 |
ENST00000379672.5
ENST00000340825.3 |
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr3_-_169587621 | 0.02 |
ENST00000523069.1
ENST00000316428.5 ENST00000264676.5 |
LRRC31
|
leucine rich repeat containing 31 |
| chr9_+_125281420 | 0.02 |
ENST00000340750.1
|
OR1J4
|
olfactory receptor, family 1, subfamily J, member 4 |
| chr6_-_131211534 | 0.02 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr4_+_20702059 | 0.02 |
ENST00000444671.2
ENST00000510700.1 ENST00000506745.1 ENST00000514663.1 ENST00000509469.1 ENST00000515339.1 ENST00000513861.1 ENST00000502374.1 ENST00000538990.1 ENST00000511160.1 ENST00000504630.1 ENST00000513590.1 ENST00000514292.1 ENST00000502938.1 ENST00000509625.1 ENST00000505160.1 ENST00000507634.1 ENST00000513459.1 ENST00000511089.1 |
PACRGL
|
PARK2 co-regulated-like |
| chr11_+_96123158 | 0.02 |
ENST00000332349.4
ENST00000458427.1 |
JRKL
|
jerky homolog-like (mouse) |
| chr10_+_43572475 | 0.02 |
ENST00000355710.3
ENST00000498820.1 ENST00000340058.5 |
RET
|
ret proto-oncogene |
| chrX_-_134305322 | 0.02 |
ENST00000276241.6
ENST00000344129.2 |
CXorf48
|
cancer/testis antigen 55 |
| chr13_-_79979952 | 0.01 |
ENST00000438724.1
|
RBM26
|
RNA binding motif protein 26 |
| chr11_+_73357614 | 0.01 |
ENST00000536527.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr4_+_81951957 | 0.01 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr3_-_122712079 | 0.01 |
ENST00000393583.2
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B |
| chr17_-_33416231 | 0.01 |
ENST00000584655.1
ENST00000447669.2 ENST00000315249.7 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr2_+_102758271 | 0.01 |
ENST00000428279.1
|
IL1R1
|
interleukin 1 receptor, type I |
| chr3_+_35721106 | 0.01 |
ENST00000474696.1
ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr18_+_5238549 | 0.01 |
ENST00000580684.1
|
LINC00667
|
long intergenic non-protein coding RNA 667 |
| chr2_-_89513402 | 0.01 |
ENST00000498435.1
|
IGKV1-27
|
immunoglobulin kappa variable 1-27 |
| chr10_-_74856608 | 0.01 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr1_+_178694362 | 0.01 |
ENST00000367634.2
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chrX_-_48216101 | 0.01 |
ENST00000298396.2
ENST00000376893.3 |
SSX3
|
synovial sarcoma, X breakpoint 3 |
| chr12_+_1929421 | 0.01 |
ENST00000543818.1
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2 |
| chr11_+_64107663 | 0.00 |
ENST00000356786.5
|
CCDC88B
|
coiled-coil domain containing 88B |
| chr7_-_93204033 | 0.00 |
ENST00000359558.2
ENST00000360249.4 ENST00000426151.1 |
CALCR
|
calcitonin receptor |
| chr20_+_43211149 | 0.00 |
ENST00000372886.1
|
PKIG
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr9_+_107266455 | 0.00 |
ENST00000334726.2
|
OR13F1
|
olfactory receptor, family 13, subfamily F, member 1 |
| chr4_-_123843597 | 0.00 |
ENST00000510735.1
ENST00000304430.5 |
NUDT6
|
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
| chrX_-_13956497 | 0.00 |
ENST00000398361.3
|
GPM6B
|
glycoprotein M6B |
| chr7_+_30323923 | 0.00 |
ENST00000323037.4
|
ZNRF2
|
zinc and ring finger 2 |
| chr12_+_120884222 | 0.00 |
ENST00000551765.1
ENST00000229384.5 |
GATC
|
glutamyl-tRNA(Gln) amidotransferase, subunit C |
Network of associatons between targets according to the STRING database.
First level regulatory network of PTF1A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.1 | 0.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 1.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) regulation of natural killer cell differentiation involved in immune response(GO:0032826) |
| 0.0 | 0.2 | GO:0032445 | fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.9 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0031627 | telomeric loop formation(GO:0031627) telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.2 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.1 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.2 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:1902728 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.6 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.6 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.7 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |