Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
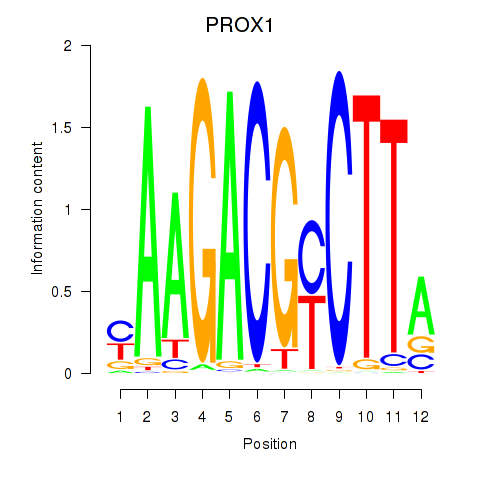
Results for PROX1
Z-value: 0.78
Transcription factors associated with PROX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PROX1
|
ENSG00000117707.11 | prospero homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PROX1 | hg19_v2_chr1_+_214161272_214161322 | 0.70 | 1.3e-01 | Click! |
Activity profile of PROX1 motif
Sorted Z-values of PROX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_142264664 | 0.47 |
ENST00000518520.1
|
RP11-10J21.3
|
Uncharacterized protein |
| chr1_+_179335101 | 0.45 |
ENST00000508285.1
ENST00000511889.1 |
AXDND1
|
axonemal dynein light chain domain containing 1 |
| chr3_-_122283424 | 0.44 |
ENST00000477522.2
ENST00000360356.2 |
PARP9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr21_+_35736302 | 0.38 |
ENST00000290310.3
|
KCNE2
|
potassium voltage-gated channel, Isk-related family, member 2 |
| chr3_+_190105909 | 0.38 |
ENST00000456423.1
|
CLDN16
|
claudin 16 |
| chr6_-_107235331 | 0.37 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr16_+_28857957 | 0.36 |
ENST00000567536.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr2_-_163175133 | 0.36 |
ENST00000421365.2
ENST00000263642.2 |
IFIH1
|
interferon induced with helicase C domain 1 |
| chr1_-_152539248 | 0.32 |
ENST00000368789.1
|
LCE3E
|
late cornified envelope 3E |
| chr3_+_122283175 | 0.30 |
ENST00000383661.3
|
DTX3L
|
deltex 3-like (Drosophila) |
| chr18_-_21562500 | 0.25 |
ENST00000582300.2
|
RP11-403A21.1
|
RP11-403A21.1 |
| chr17_+_77018896 | 0.25 |
ENST00000578229.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr21_-_16125773 | 0.25 |
ENST00000454128.2
|
AF127936.3
|
AF127936.3 |
| chr6_-_8102279 | 0.25 |
ENST00000488226.2
|
EEF1E1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr7_-_143582450 | 0.24 |
ENST00000485416.1
|
FAM115A
|
family with sequence similarity 115, member A |
| chr1_+_154966058 | 0.22 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr10_-_75385711 | 0.22 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr19_-_45909585 | 0.21 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr10_-_94301107 | 0.20 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr15_-_76304731 | 0.20 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr1_+_70876891 | 0.20 |
ENST00000411986.2
|
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr4_-_48683188 | 0.19 |
ENST00000505759.1
|
FRYL
|
FRY-like |
| chr4_-_170947565 | 0.18 |
ENST00000506764.1
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr1_+_26759295 | 0.18 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr1_+_70876926 | 0.18 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr1_+_82266053 | 0.18 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr10_+_30723045 | 0.17 |
ENST00000542547.1
ENST00000415139.1 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr10_+_134258649 | 0.17 |
ENST00000392630.3
ENST00000321248.2 |
C10orf91
|
chromosome 10 open reading frame 91 |
| chr3_-_96337000 | 0.17 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr1_-_44497118 | 0.16 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_-_107084826 | 0.16 |
ENST00000304514.7
ENST00000409886.3 |
RGPD3
|
RANBP2-like and GRIP domain containing 3 |
| chr10_+_51549498 | 0.15 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr7_-_91509986 | 0.15 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr10_-_52008313 | 0.15 |
ENST00000329428.6
ENST00000395526.4 ENST00000447815.1 |
ASAH2
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr4_+_164265035 | 0.15 |
ENST00000338566.3
|
NPY5R
|
neuropeptide Y receptor Y5 |
| chr12_-_51418549 | 0.14 |
ENST00000548150.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr2_-_122262593 | 0.14 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr13_+_49822041 | 0.14 |
ENST00000538056.1
ENST00000251108.6 ENST00000444959.1 ENST00000429346.1 |
CDADC1
|
cytidine and dCMP deaminase domain containing 1 |
| chr17_+_42081914 | 0.13 |
ENST00000293404.3
ENST00000589767.1 |
NAGS
|
N-acetylglutamate synthase |
| chr14_-_35183755 | 0.13 |
ENST00000555765.1
|
CFL2
|
cofilin 2 (muscle) |
| chr17_-_38074859 | 0.13 |
ENST00000520542.1
ENST00000418519.1 ENST00000394179.1 |
GSDMB
|
gasdermin B |
| chr9_+_133986782 | 0.13 |
ENST00000372301.2
|
AIF1L
|
allograft inflammatory factor 1-like |
| chr14_+_37641012 | 0.13 |
ENST00000556667.1
|
SLC25A21-AS1
|
SLC25A21 antisense RNA 1 |
| chr11_-_4719072 | 0.13 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr2_-_225434538 | 0.13 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chrX_-_152486108 | 0.13 |
ENST00000356661.5
|
MAGEA1
|
melanoma antigen family A, 1 (directs expression of antigen MZ2-E) |
| chr1_-_109399682 | 0.12 |
ENST00000369995.3
ENST00000370001.3 |
AKNAD1
|
AKNA domain containing 1 |
| chr10_+_54074033 | 0.12 |
ENST00000373970.3
|
DKK1
|
dickkopf WNT signaling pathway inhibitor 1 |
| chr14_-_23426231 | 0.12 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr12_+_10331605 | 0.12 |
ENST00000298530.3
|
TMEM52B
|
transmembrane protein 52B |
| chr22_-_45559540 | 0.12 |
ENST00000432502.1
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr1_-_109203685 | 0.11 |
ENST00000402983.1
ENST00000420055.1 |
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr11_+_57308979 | 0.11 |
ENST00000457912.1
|
SMTNL1
|
smoothelin-like 1 |
| chr18_+_2571510 | 0.11 |
ENST00000261597.4
ENST00000575515.1 |
NDC80
|
NDC80 kinetochore complex component |
| chr14_-_50559361 | 0.11 |
ENST00000305273.1
|
C14orf183
|
chromosome 14 open reading frame 183 |
| chr1_-_154178803 | 0.11 |
ENST00000368525.3
|
C1orf189
|
chromosome 1 open reading frame 189 |
| chr4_+_4861385 | 0.11 |
ENST00000382723.4
|
MSX1
|
msh homeobox 1 |
| chr6_-_86303833 | 0.11 |
ENST00000505648.1
|
SNX14
|
sorting nexin 14 |
| chr1_-_23342340 | 0.11 |
ENST00000566855.1
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr22_-_45559642 | 0.10 |
ENST00000426282.2
|
CTA-217C2.1
|
CTA-217C2.1 |
| chr5_-_127418573 | 0.10 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr19_-_5293243 | 0.10 |
ENST00000591760.1
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr10_+_112327425 | 0.10 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr8_+_110552831 | 0.10 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr10_+_30722866 | 0.10 |
ENST00000263056.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr19_+_45754505 | 0.10 |
ENST00000262891.4
ENST00000300843.4 |
MARK4
|
MAP/microtubule affinity-regulating kinase 4 |
| chr21_-_16126181 | 0.10 |
ENST00000455253.2
|
AF127936.3
|
AF127936.3 |
| chr3_-_33138592 | 0.10 |
ENST00000415454.1
|
GLB1
|
galactosidase, beta 1 |
| chr16_+_22518495 | 0.10 |
ENST00000541154.1
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr19_-_9546227 | 0.10 |
ENST00000361451.2
ENST00000361151.1 |
ZNF266
|
zinc finger protein 266 |
| chr12_-_46766577 | 0.10 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr19_+_53073526 | 0.10 |
ENST00000596514.1
ENST00000391785.3 ENST00000301093.2 |
ZNF701
|
zinc finger protein 701 |
| chr7_+_64254793 | 0.10 |
ENST00000494380.1
ENST00000440155.2 ENST00000440598.1 ENST00000437743.1 |
ZNF138
|
zinc finger protein 138 |
| chr3_-_130745403 | 0.10 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr16_+_28985542 | 0.10 |
ENST00000567771.1
ENST00000568388.1 |
SPNS1
|
spinster homolog 1 (Drosophila) |
| chr17_-_80009209 | 0.09 |
ENST00000429557.3
ENST00000578356.1 |
RFNG
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_-_159420780 | 0.09 |
ENST00000449822.1
|
RSPH3
|
radial spoke 3 homolog (Chlamydomonas) |
| chr16_+_58059470 | 0.09 |
ENST00000219271.3
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr3_-_25915189 | 0.09 |
ENST00000451284.1
|
LINC00692
|
long intergenic non-protein coding RNA 692 |
| chr7_+_92158083 | 0.09 |
ENST00000265732.5
ENST00000481551.1 ENST00000496410.1 |
RBM48
|
RNA binding motif protein 48 |
| chr2_+_232826394 | 0.09 |
ENST00000409401.3
ENST00000441279.1 |
DIS3L2
|
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
| chr19_-_19754404 | 0.09 |
ENST00000587205.1
ENST00000445806.2 ENST00000203556.4 |
GMIP
|
GEM interacting protein |
| chr6_+_126102292 | 0.09 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr11_+_73359936 | 0.09 |
ENST00000542389.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr13_+_113699029 | 0.09 |
ENST00000423251.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr1_+_246729724 | 0.09 |
ENST00000366513.4
ENST00000366512.3 |
CNST
|
consortin, connexin sorting protein |
| chr10_+_32735030 | 0.09 |
ENST00000277657.6
ENST00000362006.5 |
CCDC7
|
coiled-coil domain containing 7 |
| chr4_-_38666430 | 0.08 |
ENST00000436901.1
|
AC021860.1
|
Uncharacterized protein |
| chr7_+_156433363 | 0.08 |
ENST00000439609.1
|
RNF32
|
ring finger protein 32 |
| chr21_-_34863693 | 0.08 |
ENST00000314399.3
|
DNAJC28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr6_+_111580508 | 0.08 |
ENST00000368847.4
|
KIAA1919
|
KIAA1919 |
| chr11_+_6518503 | 0.08 |
ENST00000496802.1
ENST00000254579.6 ENST00000354685.3 |
DNHD1
|
dynein heavy chain domain 1 |
| chr1_-_109203648 | 0.08 |
ENST00000370031.1
|
HENMT1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr6_-_28226984 | 0.08 |
ENST00000423974.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr7_+_64254766 | 0.08 |
ENST00000307355.7
ENST00000359735.3 |
ZNF138
|
zinc finger protein 138 |
| chr7_-_91509972 | 0.08 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr2_+_163175394 | 0.08 |
ENST00000446271.1
ENST00000429691.2 |
GCA
|
grancalcin, EF-hand calcium binding protein |
| chr9_+_125132803 | 0.07 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr11_-_111749878 | 0.07 |
ENST00000260257.4
|
FDXACB1
|
ferredoxin-fold anticodon binding domain containing 1 |
| chr3_-_19975665 | 0.07 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chrY_+_22918021 | 0.07 |
ENST00000288666.5
|
RPS4Y2
|
ribosomal protein S4, Y-linked 2 |
| chr19_-_46234119 | 0.07 |
ENST00000317683.3
|
FBXO46
|
F-box protein 46 |
| chr13_+_27131887 | 0.07 |
ENST00000335327.5
|
WASF3
|
WAS protein family, member 3 |
| chr17_+_80009330 | 0.07 |
ENST00000580716.1
ENST00000583961.1 |
GPS1
|
G protein pathway suppressor 1 |
| chr4_+_26862313 | 0.07 |
ENST00000467087.1
ENST00000382009.3 ENST00000237364.5 |
STIM2
|
stromal interaction molecule 2 |
| chr4_-_170947522 | 0.07 |
ENST00000361618.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr9_+_34990219 | 0.07 |
ENST00000541010.1
ENST00000454002.2 ENST00000545841.1 |
DNAJB5
|
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr6_-_42690312 | 0.07 |
ENST00000230381.5
|
PRPH2
|
peripherin 2 (retinal degeneration, slow) |
| chr6_+_144665237 | 0.07 |
ENST00000421035.2
|
UTRN
|
utrophin |
| chr8_-_19615382 | 0.07 |
ENST00000544602.1
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr2_+_108443388 | 0.07 |
ENST00000354986.4
ENST00000408999.3 |
RGPD4
|
RANBP2-like and GRIP domain containing 4 |
| chr1_+_6640046 | 0.07 |
ENST00000319084.5
ENST00000435905.1 |
ZBTB48
|
zinc finger and BTB domain containing 48 |
| chr2_-_136288113 | 0.07 |
ENST00000401392.1
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr3_+_138067314 | 0.07 |
ENST00000423968.2
|
MRAS
|
muscle RAS oncogene homolog |
| chr19_+_6361754 | 0.07 |
ENST00000597326.1
|
CLPP
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr11_+_93394805 | 0.07 |
ENST00000325212.6
ENST00000411936.1 ENST00000344196.4 |
KIAA1731
|
KIAA1731 |
| chr3_-_130745571 | 0.07 |
ENST00000514044.1
ENST00000264992.3 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr16_-_21442874 | 0.07 |
ENST00000534903.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr2_-_65659762 | 0.07 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr4_-_128887069 | 0.06 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr14_-_54425475 | 0.06 |
ENST00000559642.1
|
BMP4
|
bone morphogenetic protein 4 |
| chr10_-_121296045 | 0.06 |
ENST00000392865.1
|
RGS10
|
regulator of G-protein signaling 10 |
| chr8_+_145064233 | 0.06 |
ENST00000529301.1
ENST00000395068.4 |
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr9_-_130497565 | 0.06 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr7_+_18126557 | 0.06 |
ENST00000417496.2
|
HDAC9
|
histone deacetylase 9 |
| chr12_+_52668394 | 0.06 |
ENST00000423955.2
|
KRT86
|
keratin 86 |
| chr11_+_73360024 | 0.06 |
ENST00000540431.1
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr9_+_95858485 | 0.06 |
ENST00000375464.2
|
C9orf89
|
chromosome 9 open reading frame 89 |
| chr11_+_117947724 | 0.06 |
ENST00000534111.1
|
TMPRSS4
|
transmembrane protease, serine 4 |
| chr8_+_145064215 | 0.06 |
ENST00000313269.5
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr7_+_100210133 | 0.06 |
ENST00000393950.2
ENST00000424091.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr10_+_16478942 | 0.05 |
ENST00000535784.2
ENST00000423462.2 ENST00000378000.1 |
PTER
|
phosphotriesterase related |
| chr3_+_126113734 | 0.05 |
ENST00000352312.1
ENST00000393425.1 |
CCDC37
|
coiled-coil domain containing 37 |
| chr8_+_110552046 | 0.05 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr2_-_175202151 | 0.05 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr5_-_22853429 | 0.05 |
ENST00000504376.2
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2) |
| chr5_+_154320623 | 0.05 |
ENST00000523037.1
ENST00000265229.8 ENST00000439747.3 ENST00000522038.1 |
MRPL22
|
mitochondrial ribosomal protein L22 |
| chr9_-_127533519 | 0.05 |
ENST00000487099.2
ENST00000344523.4 ENST00000373584.3 |
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr5_-_72861175 | 0.05 |
ENST00000504641.1
|
ANKRA2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr1_+_154909803 | 0.05 |
ENST00000604546.1
|
RP11-307C12.13
|
RP11-307C12.13 |
| chr19_+_1354930 | 0.05 |
ENST00000591337.1
|
MUM1
|
melanoma associated antigen (mutated) 1 |
| chr1_-_111061797 | 0.05 |
ENST00000369771.2
|
KCNA10
|
potassium voltage-gated channel, shaker-related subfamily, member 10 |
| chr3_+_12525931 | 0.05 |
ENST00000446004.1
ENST00000314571.7 ENST00000454502.2 ENST00000383797.5 ENST00000402228.3 ENST00000284995.6 ENST00000444864.1 |
TSEN2
|
TSEN2 tRNA splicing endonuclease subunit |
| chr15_+_85523671 | 0.05 |
ENST00000310298.4
ENST00000557957.1 |
PDE8A
|
phosphodiesterase 8A |
| chr4_-_69215699 | 0.05 |
ENST00000510746.1
ENST00000344157.4 ENST00000355665.3 |
YTHDC1
|
YTH domain containing 1 |
| chr15_+_38746307 | 0.05 |
ENST00000397609.2
ENST00000491535.1 |
FAM98B
|
family with sequence similarity 98, member B |
| chr11_+_60609537 | 0.05 |
ENST00000227520.5
|
CCDC86
|
coiled-coil domain containing 86 |
| chr16_+_23194033 | 0.05 |
ENST00000300061.2
|
SCNN1G
|
sodium channel, non-voltage-gated 1, gamma subunit |
| chr4_+_26862431 | 0.05 |
ENST00000465503.1
|
STIM2
|
stromal interaction molecule 2 |
| chr14_+_78266408 | 0.05 |
ENST00000238561.5
|
ADCK1
|
aarF domain containing kinase 1 |
| chr1_+_161719552 | 0.05 |
ENST00000367943.4
|
DUSP12
|
dual specificity phosphatase 12 |
| chr11_-_73882029 | 0.05 |
ENST00000539061.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr7_+_100209979 | 0.05 |
ENST00000493970.1
ENST00000379527.2 |
MOSPD3
|
motile sperm domain containing 3 |
| chr7_-_156433195 | 0.05 |
ENST00000333319.6
|
C7orf13
|
chromosome 7 open reading frame 13 |
| chr4_+_26862400 | 0.04 |
ENST00000467011.1
ENST00000412829.2 |
STIM2
|
stromal interaction molecule 2 |
| chr7_-_99716952 | 0.04 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr9_-_86571628 | 0.04 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr1_+_35544968 | 0.04 |
ENST00000359858.4
ENST00000373330.1 |
ZMYM1
|
zinc finger, MYM-type 1 |
| chr6_-_137540477 | 0.04 |
ENST00000367735.2
ENST00000367739.4 ENST00000458076.1 ENST00000414770.1 |
IFNGR1
|
interferon gamma receptor 1 |
| chr16_+_31724552 | 0.04 |
ENST00000539915.1
ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720
|
zinc finger protein 720 |
| chr2_+_139259324 | 0.04 |
ENST00000280098.4
|
SPOPL
|
speckle-type POZ protein-like |
| chrX_+_150565038 | 0.04 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr5_-_156569850 | 0.04 |
ENST00000524219.1
|
HAVCR2
|
hepatitis A virus cellular receptor 2 |
| chr3_+_138066539 | 0.04 |
ENST00000289104.4
|
MRAS
|
muscle RAS oncogene homolog |
| chr15_-_74726283 | 0.04 |
ENST00000543145.2
|
SEMA7A
|
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr16_-_4524543 | 0.04 |
ENST00000573571.1
ENST00000404295.3 ENST00000574425.1 |
NMRAL1
|
NmrA-like family domain containing 1 |
| chr4_+_128886584 | 0.04 |
ENST00000513371.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr14_-_105708942 | 0.04 |
ENST00000549655.1
|
BRF1
|
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
| chr9_-_127533582 | 0.04 |
ENST00000416460.2
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1 |
| chr22_+_45559722 | 0.04 |
ENST00000347635.4
ENST00000407019.2 ENST00000424634.1 ENST00000417702.1 ENST00000425733.2 ENST00000430547.1 |
NUP50
|
nucleoporin 50kDa |
| chr20_+_57209828 | 0.04 |
ENST00000598340.1
|
MGC4294
|
HCG1785561; MGC4294 protein; Uncharacterized protein |
| chr5_-_53115506 | 0.04 |
ENST00000511953.1
ENST00000504552.1 |
CTD-2081C10.1
|
CTD-2081C10.1 |
| chr20_-_7921090 | 0.04 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr13_+_27131798 | 0.04 |
ENST00000361042.4
|
WASF3
|
WAS protein family, member 3 |
| chr5_+_64859066 | 0.04 |
ENST00000261308.5
ENST00000535264.1 ENST00000538977.1 |
PPWD1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr6_-_74104856 | 0.03 |
ENST00000441145.1
|
OOEP
|
oocyte expressed protein |
| chr19_+_58281014 | 0.03 |
ENST00000391702.3
ENST00000598885.1 ENST00000598183.1 ENST00000396154.2 ENST00000599802.1 ENST00000396150.4 |
ZNF586
|
zinc finger protein 586 |
| chr1_-_204380919 | 0.03 |
ENST00000367188.4
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B |
| chr10_+_45869652 | 0.03 |
ENST00000542434.1
ENST00000374391.2 |
ALOX5
|
arachidonate 5-lipoxygenase |
| chr10_+_104474353 | 0.03 |
ENST00000602647.1
ENST00000602439.1 ENST00000602764.1 |
SFXN2
|
sideroflexin 2 |
| chr9_+_125133467 | 0.03 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr2_+_101869262 | 0.03 |
ENST00000289382.3
|
CNOT11
|
CCR4-NOT transcription complex, subunit 11 |
| chr11_+_73882144 | 0.03 |
ENST00000328257.8
|
PPME1
|
protein phosphatase methylesterase 1 |
| chr22_-_29075853 | 0.03 |
ENST00000397906.2
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr7_-_102158157 | 0.03 |
ENST00000541662.1
ENST00000306682.6 ENST00000465829.1 |
RASA4B
|
RAS p21 protein activator 4B |
| chr1_-_151431674 | 0.03 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr12_-_122658746 | 0.03 |
ENST00000377035.1
|
IL31
|
interleukin 31 |
| chr19_-_4400415 | 0.03 |
ENST00000598564.1
ENST00000417295.2 ENST00000269886.3 |
SH3GL1
|
SH3-domain GRB2-like 1 |
| chr3_-_196014520 | 0.03 |
ENST00000441879.1
ENST00000292823.2 ENST00000411591.1 ENST00000431016.1 ENST00000443555.1 |
PCYT1A
|
phosphate cytidylyltransferase 1, choline, alpha |
| chr3_-_100565249 | 0.03 |
ENST00000495591.1
ENST00000383691.4 ENST00000466947.1 |
ABI3BP
|
ABI family, member 3 (NESH) binding protein |
| chr15_+_81489213 | 0.03 |
ENST00000559383.1
ENST00000394660.2 |
IL16
|
interleukin 16 |
| chr12_-_52946923 | 0.03 |
ENST00000267119.5
|
KRT71
|
keratin 71 |
| chr8_-_146176199 | 0.03 |
ENST00000532351.1
ENST00000276816.4 ENST00000394909.2 |
ZNF16
|
zinc finger protein 16 |
| chr8_-_37824442 | 0.03 |
ENST00000345060.3
|
ADRB3
|
adrenoceptor beta 3 |
| chr3_+_137906353 | 0.03 |
ENST00000461822.1
ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr4_-_159592996 | 0.03 |
ENST00000508457.1
|
C4orf46
|
chromosome 4 open reading frame 46 |
| chr19_+_917287 | 0.03 |
ENST00000592648.1
ENST00000234371.5 |
KISS1R
|
KISS1 receptor |
| chr1_+_165864821 | 0.03 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr16_+_50300427 | 0.03 |
ENST00000394697.2
ENST00000566433.2 ENST00000538642.1 |
ADCY7
|
adenylate cyclase 7 |
| chr16_+_28858004 | 0.03 |
ENST00000322610.8
|
SH2B1
|
SH2B adaptor protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PROX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.4 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.4 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0100012 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.3 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.0 | GO:0002835 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.4 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.0 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0071893 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.0 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0001026 | TFIIIB-type transcription factor activity(GO:0001026) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |