Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
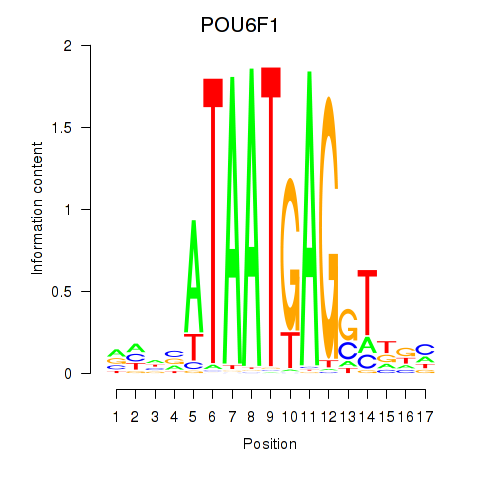
Results for POU6F1
Z-value: 0.66
Transcription factors associated with POU6F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F1
|
ENSG00000184271.11 | POU class 6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU6F1 | hg19_v2_chr12_-_51611477_51611612 | -0.47 | 3.5e-01 | Click! |
Activity profile of POU6F1 motif
Sorted Z-values of POU6F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_26746951 | 0.41 |
ENST00000523363.1
|
RP11-18A15.1
|
RP11-18A15.1 |
| chr19_+_52076425 | 0.35 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr12_+_102514019 | 0.31 |
ENST00000537257.1
ENST00000358383.5 ENST00000392911.2 |
PARPBP
|
PARP1 binding protein |
| chrX_-_122756660 | 0.30 |
ENST00000441692.1
|
THOC2
|
THO complex 2 |
| chr1_-_197115818 | 0.29 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr6_+_26183958 | 0.28 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr13_-_52027134 | 0.28 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr12_+_102513950 | 0.27 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr18_+_52495426 | 0.27 |
ENST00000262094.5
|
RAB27B
|
RAB27B, member RAS oncogene family |
| chr1_+_225140372 | 0.26 |
ENST00000366848.1
ENST00000439375.2 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr11_+_85339623 | 0.26 |
ENST00000358867.6
ENST00000534341.1 ENST00000393375.1 ENST00000531274.1 |
TMEM126B
|
transmembrane protein 126B |
| chr20_+_5987890 | 0.25 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr22_-_28490123 | 0.25 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr6_-_111804905 | 0.24 |
ENST00000358835.3
ENST00000435970.1 |
REV3L
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr2_+_207220463 | 0.23 |
ENST00000598562.1
|
AC017081.1
|
Uncharacterized protein |
| chr5_+_67586465 | 0.23 |
ENST00000336483.5
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chrY_+_14958970 | 0.22 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr7_-_83824449 | 0.22 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr17_-_25568687 | 0.20 |
ENST00000581944.1
|
RP11-663N22.1
|
RP11-663N22.1 |
| chr6_-_47009996 | 0.20 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chr1_+_225117350 | 0.20 |
ENST00000413949.2
ENST00000430092.1 ENST00000366850.3 ENST00000400952.3 ENST00000366849.1 |
DNAH14
|
dynein, axonemal, heavy chain 14 |
| chr15_-_85259384 | 0.18 |
ENST00000455959.3
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr2_+_242289502 | 0.18 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr4_+_78829479 | 0.17 |
ENST00000504901.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr2_+_191045562 | 0.17 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr15_+_80351977 | 0.17 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr13_+_98612446 | 0.17 |
ENST00000496368.1
ENST00000421861.2 ENST00000357602.3 |
IPO5
|
importin 5 |
| chr5_+_162864575 | 0.16 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr20_-_50418947 | 0.15 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr4_-_169401628 | 0.15 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chrX_+_119737806 | 0.14 |
ENST00000371317.5
|
MCTS1
|
malignant T cell amplified sequence 1 |
| chr10_-_94257512 | 0.14 |
ENST00000371581.5
|
IDE
|
insulin-degrading enzyme |
| chr5_+_115177178 | 0.13 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr15_-_85259294 | 0.13 |
ENST00000558217.1
ENST00000558196.1 ENST00000558134.1 |
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr14_+_39944025 | 0.13 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr4_-_130692631 | 0.12 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr14_+_24970800 | 0.12 |
ENST00000555109.1
|
RP11-80A15.1
|
Uncharacterized protein |
| chr6_-_110501126 | 0.12 |
ENST00000368938.1
|
WASF1
|
WAS protein family, member 1 |
| chr3_-_108248169 | 0.12 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr13_+_107028897 | 0.12 |
ENST00000439790.1
ENST00000435024.1 |
LINC00460
|
long intergenic non-protein coding RNA 460 |
| chr22_+_31518938 | 0.11 |
ENST00000412985.1
ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J
|
inositol polyphosphate-5-phosphatase J |
| chr3_-_114343768 | 0.11 |
ENST00000393785.2
|
ZBTB20
|
zinc finger and BTB domain containing 20 |
| chr2_+_128403720 | 0.11 |
ENST00000272644.3
|
GPR17
|
G protein-coupled receptor 17 |
| chr1_-_153538292 | 0.11 |
ENST00000497140.1
ENST00000368708.3 |
S100A2
|
S100 calcium binding protein A2 |
| chrX_+_102024075 | 0.11 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr13_+_49551020 | 0.10 |
ENST00000541916.1
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chr11_+_12302492 | 0.10 |
ENST00000533534.1
|
MICALCL
|
MICAL C-terminal like |
| chr15_+_80351910 | 0.10 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr7_-_83824169 | 0.09 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr6_-_111136299 | 0.09 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr12_-_95397442 | 0.09 |
ENST00000547157.1
ENST00000547986.1 ENST00000327772.2 |
NDUFA12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr6_+_110501344 | 0.08 |
ENST00000368932.1
|
CDC40
|
cell division cycle 40 |
| chr6_+_110501621 | 0.08 |
ENST00000368930.1
ENST00000307731.1 |
CDC40
|
cell division cycle 40 |
| chr7_-_77427676 | 0.08 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr2_+_102721023 | 0.08 |
ENST00000409589.1
ENST00000409329.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr4_-_83812157 | 0.08 |
ENST00000513323.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr15_-_85259330 | 0.07 |
ENST00000560266.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr11_+_118398178 | 0.07 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chr4_+_123161120 | 0.07 |
ENST00000446180.1
|
KIAA1109
|
KIAA1109 |
| chr12_-_57443886 | 0.07 |
ENST00000300119.3
|
MYO1A
|
myosin IA |
| chr4_-_76912070 | 0.07 |
ENST00000395711.4
ENST00000356260.5 |
SDAD1
|
SDA1 domain containing 1 |
| chr2_+_128403439 | 0.07 |
ENST00000544369.1
|
GPR17
|
G protein-coupled receptor 17 |
| chr19_+_10959043 | 0.06 |
ENST00000397820.4
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr11_-_107729287 | 0.06 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr9_+_128509624 | 0.06 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr1_-_106161540 | 0.05 |
ENST00000420901.1
ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1
|
RP11-251P6.1 |
| chr11_-_107729504 | 0.05 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr15_-_31283618 | 0.05 |
ENST00000563714.1
|
MTMR10
|
myotubularin related protein 10 |
| chr20_+_58515417 | 0.05 |
ENST00000360816.3
|
FAM217B
|
family with sequence similarity 217, member B |
| chr6_-_74231444 | 0.04 |
ENST00000331523.2
ENST00000356303.2 |
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr9_+_103790991 | 0.04 |
ENST00000374874.3
|
LPPR1
|
Lipid phosphate phosphatase-related protein type 1 |
| chr4_+_130692778 | 0.04 |
ENST00000513875.1
ENST00000508724.1 |
RP11-422J15.1
|
RP11-422J15.1 |
| chr12_-_91546926 | 0.04 |
ENST00000550758.1
|
DCN
|
decorin |
| chr10_+_97803151 | 0.03 |
ENST00000403870.3
ENST00000265992.5 ENST00000465148.2 ENST00000534974.1 |
CCNJ
|
cyclin J |
| chr9_-_4666337 | 0.03 |
ENST00000381890.5
|
SPATA6L
|
spermatogenesis associated 6-like |
| chr12_+_49297887 | 0.03 |
ENST00000266984.5
|
CCDC65
|
coiled-coil domain containing 65 |
| chr16_+_19421803 | 0.03 |
ENST00000541464.1
|
TMC5
|
transmembrane channel-like 5 |
| chr15_-_44828838 | 0.03 |
ENST00000560750.1
|
EIF3J-AS1
|
EIF3J antisense RNA 1 (head to head) |
| chrX_+_43515467 | 0.03 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr8_-_10512569 | 0.03 |
ENST00000382483.3
|
RP1L1
|
retinitis pigmentosa 1-like 1 |
| chr1_+_113010056 | 0.03 |
ENST00000369686.5
|
WNT2B
|
wingless-type MMTV integration site family, member 2B |
| chr11_+_2323349 | 0.03 |
ENST00000381121.3
|
TSPAN32
|
tetraspanin 32 |
| chr15_-_85259360 | 0.03 |
ENST00000559729.1
|
SEC11A
|
SEC11 homolog A (S. cerevisiae) |
| chr11_-_93583697 | 0.02 |
ENST00000409977.1
|
VSTM5
|
V-set and transmembrane domain containing 5 |
| chr6_-_111136513 | 0.02 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr12_+_10460417 | 0.02 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr17_-_39677971 | 0.02 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr2_+_202937972 | 0.02 |
ENST00000541917.1
ENST00000295844.3 |
AC079354.1
|
uncharacterized protein KIAA2012 |
| chr20_+_58203664 | 0.02 |
ENST00000541461.1
|
PHACTR3
|
phosphatase and actin regulator 3 |
| chr11_+_2323236 | 0.02 |
ENST00000182290.4
|
TSPAN32
|
tetraspanin 32 |
| chr12_+_54892550 | 0.01 |
ENST00000545638.2
|
NCKAP1L
|
NCK-associated protein 1-like |
| chr15_+_45028753 | 0.01 |
ENST00000338264.4
|
TRIM69
|
tripartite motif containing 69 |
| chr19_-_3786354 | 0.01 |
ENST00000395040.2
ENST00000310132.6 |
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr9_-_13165457 | 0.01 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr15_+_45028520 | 0.01 |
ENST00000329464.4
|
TRIM69
|
tripartite motif containing 69 |
| chr10_+_18041218 | 0.01 |
ENST00000480516.1
ENST00000457860.1 |
TMEM236
|
TMEM236 |
| chr20_-_58515344 | 0.00 |
ENST00000370996.3
|
PPP1R3D
|
protein phosphatase 1, regulatory subunit 3D |
| chr20_-_50418972 | 0.00 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr16_+_19422035 | 0.00 |
ENST00000381414.4
ENST00000396229.2 |
TMC5
|
transmembrane channel-like 5 |
| chr8_+_39792474 | 0.00 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr19_-_3786253 | 0.00 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr9_-_89562104 | 0.00 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU6F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.3 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.6 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.3 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.1 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.0 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.2 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030572 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |