Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
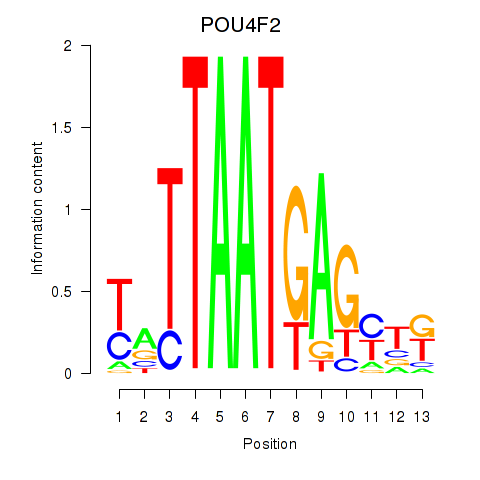
Results for POU4F2
Z-value: 0.50
Transcription factors associated with POU4F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F2
|
ENSG00000151615.3 | POU class 4 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU4F2 | hg19_v2_chr4_+_147560042_147560046 | 0.32 | 5.3e-01 | Click! |
Activity profile of POU4F2 motif
Sorted Z-values of POU4F2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_51027838 | 0.34 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr4_+_141264597 | 0.30 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr19_-_7698599 | 0.30 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr6_-_111136299 | 0.28 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr9_-_37384431 | 0.28 |
ENST00000452923.1
|
RP11-397D12.4
|
RP11-397D12.4 |
| chr5_-_87516448 | 0.22 |
ENST00000511218.1
|
TMEM161B
|
transmembrane protein 161B |
| chr17_+_61151306 | 0.20 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr12_-_118796910 | 0.19 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr18_-_73967160 | 0.17 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr13_-_34250861 | 0.16 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chrX_+_139791917 | 0.16 |
ENST00000607004.1
ENST00000370535.3 |
LINC00632
|
long intergenic non-protein coding RNA 632 |
| chr6_+_130339710 | 0.14 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr6_-_111136513 | 0.14 |
ENST00000368911.3
|
CDK19
|
cyclin-dependent kinase 19 |
| chr11_-_125550764 | 0.13 |
ENST00000527795.1
|
ACRV1
|
acrosomal vesicle protein 1 |
| chr20_+_5987890 | 0.12 |
ENST00000378868.4
|
CRLS1
|
cardiolipin synthase 1 |
| chr11_-_125550726 | 0.12 |
ENST00000315608.3
ENST00000530048.1 |
ACRV1
|
acrosomal vesicle protein 1 |
| chr2_+_191792376 | 0.12 |
ENST00000409428.1
ENST00000409215.1 |
GLS
|
glutaminase |
| chr8_-_18541603 | 0.12 |
ENST00000428502.2
|
PSD3
|
pleckstrin and Sec7 domain containing 3 |
| chr15_+_45544426 | 0.11 |
ENST00000347644.3
ENST00000560438.1 |
SLC28A2
|
solute carrier family 28 (concentrative nucleoside transporter), member 2 |
| chr1_-_193028632 | 0.11 |
ENST00000421683.1
|
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr11_+_31531291 | 0.11 |
ENST00000350638.5
ENST00000379163.5 ENST00000395934.2 |
ELP4
|
elongator acetyltransferase complex subunit 4 |
| chr19_-_7697857 | 0.10 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
| chr2_+_44502597 | 0.10 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr14_+_55221541 | 0.10 |
ENST00000555192.1
|
SAMD4A
|
sterile alpha motif domain containing 4A |
| chr8_-_131399110 | 0.09 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr2_+_172309634 | 0.09 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr17_+_48243352 | 0.09 |
ENST00000344627.6
ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA
|
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr4_-_36245561 | 0.09 |
ENST00000506189.1
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr4_-_169401628 | 0.09 |
ENST00000514748.1
ENST00000512371.1 ENST00000260184.7 ENST00000505890.1 ENST00000511577.1 |
DDX60L
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
| chr1_-_193028426 | 0.08 |
ENST00000367450.3
ENST00000530098.2 ENST00000367451.4 ENST00000367448.1 ENST00000367449.1 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr4_+_169013666 | 0.08 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr9_+_125132803 | 0.08 |
ENST00000540753.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr15_-_37393406 | 0.08 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr4_+_76481258 | 0.08 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr1_-_193028621 | 0.07 |
ENST00000367455.4
ENST00000367454.1 |
UCHL5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr16_-_69390209 | 0.07 |
ENST00000563634.1
|
RP11-343C2.9
|
Uncharacterized protein |
| chr12_-_89746173 | 0.07 |
ENST00000308385.6
|
DUSP6
|
dual specificity phosphatase 6 |
| chr15_+_101402041 | 0.07 |
ENST00000558475.1
ENST00000558641.1 ENST00000559673.1 |
RP11-66B24.1
|
RP11-66B24.1 |
| chr21_+_17443521 | 0.07 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_193028552 | 0.06 |
ENST00000400968.2
ENST00000432079.1 |
TROVE2
|
TROVE domain family, member 2 |
| chr6_+_12717892 | 0.06 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr17_-_64225508 | 0.06 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chrX_+_133930798 | 0.06 |
ENST00000414371.2
|
FAM122C
|
family with sequence similarity 122C |
| chr20_-_50418972 | 0.05 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr17_+_68100989 | 0.05 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr9_+_125133315 | 0.05 |
ENST00000223423.4
ENST00000362012.2 |
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr11_-_107729287 | 0.05 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr7_+_142982023 | 0.05 |
ENST00000359333.3
ENST00000409244.1 ENST00000409541.1 ENST00000410004.1 |
TMEM139
|
transmembrane protein 139 |
| chr1_+_160370344 | 0.05 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chrX_+_68835911 | 0.05 |
ENST00000525810.1
ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA
|
ectodysplasin A |
| chr12_-_130529501 | 0.05 |
ENST00000561864.1
ENST00000567788.1 |
RP11-474D1.4
RP11-474D1.3
|
RP11-474D1.4 RP11-474D1.3 |
| chr10_-_73848086 | 0.05 |
ENST00000536168.1
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr11_-_96076334 | 0.05 |
ENST00000524717.1
|
MAML2
|
mastermind-like 2 (Drosophila) |
| chr13_+_46039037 | 0.04 |
ENST00000349995.5
|
COG3
|
component of oligomeric golgi complex 3 |
| chr12_-_52779433 | 0.04 |
ENST00000257951.3
|
KRT84
|
keratin 84 |
| chr11_-_31531121 | 0.04 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr10_-_73848531 | 0.04 |
ENST00000373109.2
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr10_-_116444371 | 0.04 |
ENST00000533213.2
ENST00000369252.4 |
ABLIM1
|
actin binding LIM protein 1 |
| chr2_+_44502630 | 0.04 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr13_-_74993252 | 0.04 |
ENST00000325811.1
|
AL355390.1
|
Uncharacterized protein |
| chr6_+_4087664 | 0.04 |
ENST00000430835.2
|
C6orf201
|
chromosome 6 open reading frame 201 |
| chr9_-_79520989 | 0.04 |
ENST00000376713.3
ENST00000376718.3 ENST00000428286.1 |
PRUNE2
|
prune homolog 2 (Drosophila) |
| chr6_+_151042224 | 0.03 |
ENST00000358517.2
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_+_198608146 | 0.03 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr1_+_193028717 | 0.03 |
ENST00000415442.2
ENST00000506303.1 |
TROVE2
|
TROVE domain family, member 2 |
| chr19_+_1269324 | 0.02 |
ENST00000589710.1
ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP
|
cold inducible RNA binding protein |
| chr2_-_122262593 | 0.02 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr9_-_5830768 | 0.02 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr2_-_158300556 | 0.02 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr21_+_17442799 | 0.02 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr3_+_147657764 | 0.02 |
ENST00000467198.1
ENST00000485006.1 |
RP11-71N10.1
|
RP11-71N10.1 |
| chr1_+_66820058 | 0.02 |
ENST00000480109.2
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr20_+_56136136 | 0.02 |
ENST00000319441.4
ENST00000543666.1 |
PCK1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr12_-_118797475 | 0.02 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr3_+_50230105 | 0.02 |
ENST00000440836.1
|
GNAT1
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 |
| chr9_+_125133467 | 0.02 |
ENST00000426608.1
|
PTGS1
|
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr19_-_36304201 | 0.02 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr18_-_71959159 | 0.02 |
ENST00000494131.2
ENST00000397914.4 ENST00000340533.4 |
CYB5A
|
cytochrome b5 type A (microsomal) |
| chr6_+_39760129 | 0.02 |
ENST00000274867.4
|
DAAM2
|
dishevelled associated activator of morphogenesis 2 |
| chr5_-_58295712 | 0.01 |
ENST00000317118.8
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr16_+_19422035 | 0.01 |
ENST00000381414.4
ENST00000396229.2 |
TMC5
|
transmembrane channel-like 5 |
| chrX_-_133931164 | 0.01 |
ENST00000370790.1
ENST00000298090.6 |
FAM122B
|
family with sequence similarity 122B |
| chr10_+_95372289 | 0.01 |
ENST00000371447.3
|
PDE6C
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime |
| chr12_-_89746264 | 0.01 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr12_+_109577202 | 0.00 |
ENST00000377848.3
ENST00000377854.5 |
ACACB
|
acetyl-CoA carboxylase beta |
| chr11_-_115127611 | 0.00 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr12_-_6233828 | 0.00 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr17_-_33390667 | 0.00 |
ENST00000378516.2
ENST00000268850.7 ENST00000394597.2 |
RFFL
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr5_+_154393260 | 0.00 |
ENST00000435029.4
|
KIF4B
|
kinesin family member 4B |
| chr10_-_99205607 | 0.00 |
ENST00000477692.2
ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1
|
exosome component 1 |
| chr18_+_32455201 | 0.00 |
ENST00000590831.2
|
DTNA
|
dystrobrevin, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU4F2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.0 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.3 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |