Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
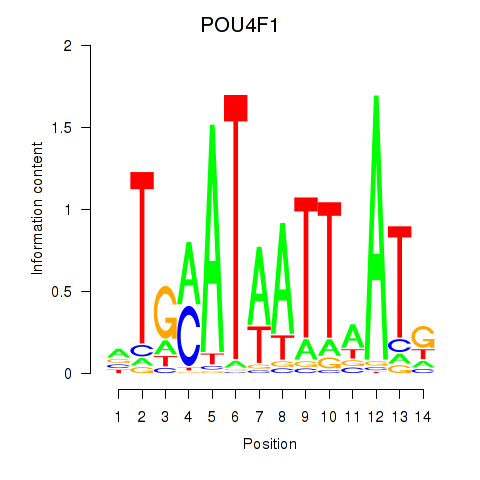
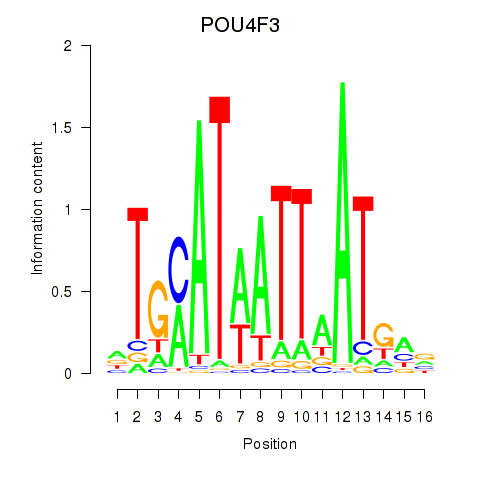
Results for POU4F1_POU4F3
Z-value: 0.74
Transcription factors associated with POU4F1_POU4F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F1
|
ENSG00000152192.6 | POU class 4 homeobox 1 |
|
POU4F3
|
ENSG00000091010.4 | POU class 4 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU4F1 | hg19_v2_chr13_-_79177673_79177701 | 0.34 | 5.0e-01 | Click! |
| POU4F3 | hg19_v2_chr5_+_145718587_145718607 | 0.12 | 8.3e-01 | Click! |
Activity profile of POU4F1_POU4F3 motif
Sorted Z-values of POU4F1_POU4F3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_121624973 | 0.85 |
ENST00000603720.1
|
RP11-297J22.1
|
RP11-297J22.1 |
| chr12_-_58212487 | 0.71 |
ENST00000549994.1
|
AVIL
|
advillin |
| chrX_-_106146547 | 0.59 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr22_+_39101728 | 0.52 |
ENST00000216044.5
ENST00000484657.1 |
GTPBP1
|
GTP binding protein 1 |
| chr11_-_58378759 | 0.51 |
ENST00000601906.1
|
AP001350.1
|
Uncharacterized protein |
| chrX_+_47053208 | 0.51 |
ENST00000442035.1
ENST00000457753.1 ENST00000335972.6 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr17_+_61151306 | 0.46 |
ENST00000580068.1
ENST00000580466.1 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chrX_+_47050798 | 0.44 |
ENST00000412206.1
ENST00000427561.1 |
UBA1
|
ubiquitin-like modifier activating enzyme 1 |
| chr19_+_48949087 | 0.44 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr6_-_26108355 | 0.43 |
ENST00000338379.4
|
HIST1H1T
|
histone cluster 1, H1t |
| chr16_-_4164027 | 0.38 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
| chr4_-_89442940 | 0.38 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr22_-_39268308 | 0.36 |
ENST00000407418.3
|
CBX6
|
chromobox homolog 6 |
| chr7_+_100026406 | 0.34 |
ENST00000414441.1
|
MEPCE
|
methylphosphate capping enzyme |
| chr8_+_98900132 | 0.32 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr19_-_55677999 | 0.31 |
ENST00000532817.1
ENST00000527223.2 ENST00000391720.4 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr19_+_48949030 | 0.30 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr19_+_39138320 | 0.29 |
ENST00000424234.2
ENST00000390009.3 ENST00000589528.1 |
ACTN4
|
actinin, alpha 4 |
| chr15_+_76352178 | 0.28 |
ENST00000388942.3
|
C15orf27
|
chromosome 15 open reading frame 27 |
| chr12_-_6798616 | 0.27 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr1_+_81001398 | 0.26 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chrX_-_39956656 | 0.26 |
ENST00000397354.3
ENST00000378444.4 |
BCOR
|
BCL6 corepressor |
| chr19_+_39138271 | 0.25 |
ENST00000252699.2
|
ACTN4
|
actinin, alpha 4 |
| chrX_-_1331527 | 0.25 |
ENST00000381567.3
ENST00000381566.1 ENST00000400841.2 |
CRLF2
|
cytokine receptor-like factor 2 |
| chr7_+_150725510 | 0.23 |
ENST00000461373.1
ENST00000358849.4 ENST00000297504.6 ENST00000542328.1 ENST00000498578.1 ENST00000356058.4 ENST00000477719.1 ENST00000477092.1 |
ABCB8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr15_+_58724184 | 0.21 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr18_+_29027696 | 0.21 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr22_-_43010928 | 0.21 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr2_-_101034070 | 0.21 |
ENST00000264249.3
|
CHST10
|
carbohydrate sulfotransferase 10 |
| chr12_+_51984657 | 0.21 |
ENST00000550891.1
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr1_+_149239529 | 0.21 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr12_-_6798410 | 0.21 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr12_-_6798523 | 0.21 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr9_-_34458531 | 0.20 |
ENST00000379089.1
ENST00000379087.1 ENST00000379084.1 ENST00000379081.1 ENST00000379080.1 ENST00000422409.1 ENST00000379078.1 ENST00000445726.1 ENST00000297620.4 |
FAM219A
|
family with sequence similarity 219, member A |
| chr14_+_101297740 | 0.20 |
ENST00000555928.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr1_+_160336851 | 0.20 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr16_-_70835034 | 0.19 |
ENST00000261776.5
|
VAC14
|
Vac14 homolog (S. cerevisiae) |
| chr12_+_110011571 | 0.19 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr6_-_116833500 | 0.19 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr6_-_127840336 | 0.18 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr1_+_145292806 | 0.18 |
ENST00000448873.2
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr10_+_135207598 | 0.17 |
ENST00000477902.2
|
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr6_-_127840048 | 0.17 |
ENST00000467753.1
|
SOGA3
|
SOGA family member 3 |
| chr17_+_17685422 | 0.17 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr22_+_43011247 | 0.16 |
ENST00000602478.1
|
RNU12
|
RNA, U12 small nuclear |
| chr6_-_127840021 | 0.16 |
ENST00000465909.2
|
SOGA3
|
SOGA family member 3 |
| chr19_-_55677920 | 0.16 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr10_+_135207623 | 0.16 |
ENST00000317502.6
ENST00000432508.3 |
MTG1
|
mitochondrial ribosome-associated GTPase 1 |
| chr8_-_125577940 | 0.16 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr3_-_155011483 | 0.16 |
ENST00000489090.1
|
RP11-451G4.2
|
RP11-451G4.2 |
| chr22_-_43010859 | 0.16 |
ENST00000339677.6
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr15_-_42264702 | 0.15 |
ENST00000220325.4
|
EHD4
|
EH-domain containing 4 |
| chr3_+_35722487 | 0.15 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr10_+_695888 | 0.15 |
ENST00000441152.2
|
PRR26
|
proline rich 26 |
| chr1_+_81106951 | 0.15 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr10_+_81891416 | 0.15 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr10_-_75226166 | 0.15 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr20_-_33872548 | 0.15 |
ENST00000374443.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr15_-_54267147 | 0.14 |
ENST00000558866.1
ENST00000558920.1 |
RP11-643A5.2
|
RP11-643A5.2 |
| chr7_+_134528635 | 0.14 |
ENST00000445569.2
|
CALD1
|
caldesmon 1 |
| chr1_-_155214436 | 0.14 |
ENST00000327247.5
|
GBA
|
glucosidase, beta, acid |
| chr12_-_120241187 | 0.14 |
ENST00000392520.2
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr5_+_42756903 | 0.14 |
ENST00000361970.5
ENST00000388827.4 |
CCDC152
|
coiled-coil domain containing 152 |
| chr7_-_100026280 | 0.14 |
ENST00000360951.4
ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1
|
zinc finger, CW type with PWWP domain 1 |
| chr6_-_152623231 | 0.13 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_160549235 | 0.13 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr20_+_30102231 | 0.13 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chr10_+_118349920 | 0.13 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr16_+_619931 | 0.13 |
ENST00000321878.5
ENST00000439574.1 ENST00000026218.5 ENST00000470411.2 |
PIGQ
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr9_+_131683174 | 0.12 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr12_-_51611477 | 0.12 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr22_-_30970560 | 0.12 |
ENST00000402369.1
ENST00000406361.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr19_-_53696587 | 0.12 |
ENST00000396424.3
ENST00000600412.1 |
ZNF665
|
zinc finger protein 665 |
| chr3_+_35722424 | 0.12 |
ENST00000396481.2
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr6_-_44281043 | 0.12 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr6_+_30856507 | 0.12 |
ENST00000513240.1
ENST00000424544.2 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_-_26593649 | 0.12 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr18_-_52989217 | 0.11 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr6_-_136847610 | 0.11 |
ENST00000454590.1
ENST00000432797.2 |
MAP7
|
microtubule-associated protein 7 |
| chr3_+_158787041 | 0.11 |
ENST00000471575.1
ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1
|
IQCJ-SCHIP1 readthrough |
| chr16_+_15596123 | 0.11 |
ENST00000452191.2
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr3_+_167582561 | 0.11 |
ENST00000463642.1
ENST00000464514.1 |
RP11-298O21.6
RP11-298O21.7
|
RP11-298O21.6 RP11-298O21.7 |
| chr5_-_135290705 | 0.11 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr1_-_156470515 | 0.11 |
ENST00000340875.5
ENST00000368240.2 ENST00000353795.3 |
MEF2D
|
myocyte enhancer factor 2D |
| chrX_+_10126488 | 0.11 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr10_-_6104253 | 0.10 |
ENST00000256876.6
ENST00000379954.1 ENST00000379959.3 |
IL2RA
|
interleukin 2 receptor, alpha |
| chr1_-_155214475 | 0.10 |
ENST00000428024.3
|
GBA
|
glucosidase, beta, acid |
| chr2_-_88285309 | 0.10 |
ENST00000420840.2
|
RGPD2
|
RANBP2-like and GRIP domain containing 2 |
| chr12_-_89746264 | 0.10 |
ENST00000548755.1
|
DUSP6
|
dual specificity phosphatase 6 |
| chr11_+_36317830 | 0.10 |
ENST00000530639.1
|
PRR5L
|
proline rich 5 like |
| chr17_+_67590125 | 0.10 |
ENST00000591334.1
|
AC003051.1
|
AC003051.1 |
| chr12_+_109785708 | 0.10 |
ENST00000310903.5
|
MYO1H
|
myosin IH |
| chr11_-_111649015 | 0.10 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr5_+_157158205 | 0.10 |
ENST00000231198.7
|
THG1L
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr10_-_22292675 | 0.10 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr6_-_2634820 | 0.10 |
ENST00000296847.3
|
C6orf195
|
chromosome 6 open reading frame 195 |
| chr20_+_36405665 | 0.10 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr18_+_61575200 | 0.10 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr1_+_165864800 | 0.09 |
ENST00000469256.2
|
UCK2
|
uridine-cytidine kinase 2 |
| chr6_+_12717892 | 0.09 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr19_+_52873166 | 0.09 |
ENST00000424032.2
ENST00000600321.1 ENST00000344085.5 ENST00000597976.1 ENST00000422689.2 |
ZNF880
|
zinc finger protein 880 |
| chr2_+_179318295 | 0.09 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr1_-_197036364 | 0.09 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr7_-_5465045 | 0.09 |
ENST00000399434.2
|
TNRC18
|
trinucleotide repeat containing 18 |
| chr3_+_35722844 | 0.09 |
ENST00000436702.1
ENST00000438071.1 |
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr1_-_33430286 | 0.09 |
ENST00000373456.7
ENST00000356990.5 ENST00000235150.4 |
RNF19B
|
ring finger protein 19B |
| chr1_-_36020531 | 0.09 |
ENST00000440579.1
ENST00000494948.1 |
KIAA0319L
|
KIAA0319-like |
| chr2_+_133874577 | 0.08 |
ENST00000596384.1
|
AC011755.1
|
HCG2006742; Protein LOC100996685 |
| chr22_-_30970498 | 0.08 |
ENST00000431313.1
|
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr16_-_67217844 | 0.08 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr9_+_135906076 | 0.08 |
ENST00000372097.5
ENST00000440319.1 |
GTF3C5
|
general transcription factor IIIC, polypeptide 5, 63kDa |
| chr11_+_44117099 | 0.08 |
ENST00000533608.1
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr12_-_11139511 | 0.08 |
ENST00000506868.1
|
TAS2R50
|
taste receptor, type 2, member 50 |
| chr21_+_40181520 | 0.08 |
ENST00000456966.1
|
ETS2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr1_+_67773044 | 0.08 |
ENST00000262345.1
ENST00000371000.1 |
IL12RB2
|
interleukin 12 receptor, beta 2 |
| chr12_-_111926342 | 0.08 |
ENST00000389154.3
|
ATXN2
|
ataxin 2 |
| chr11_-_559377 | 0.08 |
ENST00000486629.1
|
C11orf35
|
chromosome 11 open reading frame 35 |
| chr1_+_205197304 | 0.08 |
ENST00000358024.3
|
TMCC2
|
transmembrane and coiled-coil domain family 2 |
| chr2_-_55237484 | 0.08 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr21_-_37914898 | 0.08 |
ENST00000399136.1
|
CLDN14
|
claudin 14 |
| chr6_-_136847099 | 0.08 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr12_+_51985001 | 0.07 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr1_+_165864821 | 0.07 |
ENST00000470820.1
|
UCK2
|
uridine-cytidine kinase 2 |
| chr17_+_7155819 | 0.07 |
ENST00000570322.1
ENST00000576496.1 ENST00000574841.2 |
ELP5
|
elongator acetyltransferase complex subunit 5 |
| chr2_-_3504587 | 0.07 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr14_-_65409438 | 0.07 |
ENST00000557049.1
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr6_-_161695074 | 0.07 |
ENST00000457520.2
ENST00000366906.5 ENST00000320285.4 |
AGPAT4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr12_-_49582978 | 0.07 |
ENST00000301071.7
|
TUBA1A
|
tubulin, alpha 1a |
| chr10_+_104614008 | 0.07 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr19_+_16940198 | 0.07 |
ENST00000248054.5
ENST00000596802.1 ENST00000379803.1 |
SIN3B
|
SIN3 transcription regulator family member B |
| chr13_+_24144796 | 0.07 |
ENST00000403372.2
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr6_-_127840453 | 0.07 |
ENST00000556132.1
|
SOGA3
|
SOGA family member 3 |
| chr14_-_65409502 | 0.07 |
ENST00000389614.5
|
GPX2
|
glutathione peroxidase 2 (gastrointestinal) |
| chr10_-_128359074 | 0.07 |
ENST00000544758.1
|
C10orf90
|
chromosome 10 open reading frame 90 |
| chr2_+_65663812 | 0.06 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr16_-_51185149 | 0.06 |
ENST00000566102.1
ENST00000541611.1 |
SALL1
|
spalt-like transcription factor 1 |
| chr1_+_73771844 | 0.06 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr4_-_122085469 | 0.06 |
ENST00000057513.3
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr6_-_76782371 | 0.06 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chr19_+_54466179 | 0.06 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr11_+_44117219 | 0.06 |
ENST00000532479.1
ENST00000527014.1 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr10_+_696000 | 0.06 |
ENST00000381489.5
|
PRR26
|
proline rich 26 |
| chr10_-_419129 | 0.06 |
ENST00000540204.1
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr16_-_14109841 | 0.06 |
ENST00000576797.1
ENST00000575424.1 |
CTD-2135D7.5
|
CTD-2135D7.5 |
| chr22_+_41777927 | 0.06 |
ENST00000266304.4
|
TEF
|
thyrotrophic embryonic factor |
| chr9_-_95298254 | 0.06 |
ENST00000444490.2
|
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr10_-_116418053 | 0.06 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr19_-_18632861 | 0.06 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr11_-_8290263 | 0.06 |
ENST00000428101.2
|
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr12_-_122879969 | 0.06 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr2_+_87135076 | 0.05 |
ENST00000409776.2
|
RGPD1
|
RANBP2-like and GRIP domain containing 1 |
| chr14_+_31959154 | 0.05 |
ENST00000550005.1
|
NUBPL
|
nucleotide binding protein-like |
| chr8_-_52721975 | 0.05 |
ENST00000356297.4
ENST00000543296.1 |
PXDNL
|
peroxidasin homolog (Drosophila)-like |
| chr20_-_1447467 | 0.05 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr6_-_146057144 | 0.05 |
ENST00000367519.3
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chrX_+_41548259 | 0.05 |
ENST00000378138.5
|
GPR34
|
G protein-coupled receptor 34 |
| chr18_-_70305745 | 0.05 |
ENST00000581073.1
|
CBLN2
|
cerebellin 2 precursor |
| chr19_-_20748541 | 0.05 |
ENST00000427401.4
ENST00000594419.1 |
ZNF737
|
zinc finger protein 737 |
| chr1_+_145293114 | 0.05 |
ENST00000369338.1
|
NBPF10
|
neuroblastoma breakpoint family, member 10 |
| chr12_-_49582837 | 0.05 |
ENST00000547939.1
ENST00000546918.1 ENST00000552924.1 |
TUBA1A
|
tubulin, alpha 1a |
| chr7_-_115670792 | 0.05 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr6_-_27880174 | 0.05 |
ENST00000303324.2
|
OR2B2
|
olfactory receptor, family 2, subfamily B, member 2 |
| chr17_-_72855989 | 0.05 |
ENST00000293190.5
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
| chr20_-_1447547 | 0.05 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr11_-_102709441 | 0.05 |
ENST00000434103.1
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr2_-_183106641 | 0.05 |
ENST00000346717.4
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr22_+_31488433 | 0.05 |
ENST00000455608.1
|
SMTN
|
smoothelin |
| chr17_-_49124230 | 0.04 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr4_-_69817481 | 0.04 |
ENST00000251566.4
|
UGT2A3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr5_-_77844974 | 0.04 |
ENST00000515007.2
|
LHFPL2
|
lipoma HMGIC fusion partner-like 2 |
| chr3_+_70048881 | 0.04 |
ENST00000483525.1
|
RP11-460N16.1
|
RP11-460N16.1 |
| chr12_+_861717 | 0.04 |
ENST00000535572.1
|
WNK1
|
WNK lysine deficient protein kinase 1 |
| chrX_-_15332665 | 0.04 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_+_133174147 | 0.04 |
ENST00000329321.3
|
GPR39
|
G protein-coupled receptor 39 |
| chr6_-_33297013 | 0.04 |
ENST00000453407.1
|
DAXX
|
death-domain associated protein |
| chr6_+_34204642 | 0.04 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr2_-_70475730 | 0.04 |
ENST00000445587.1
ENST00000433529.2 ENST00000415783.2 |
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr17_-_56492989 | 0.04 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr16_+_69221028 | 0.04 |
ENST00000336278.4
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr10_-_11574274 | 0.04 |
ENST00000277575.5
|
USP6NL
|
USP6 N-terminal like |
| chr17_-_40540484 | 0.04 |
ENST00000588969.1
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr14_+_39703112 | 0.04 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr1_+_161691353 | 0.04 |
ENST00000367948.2
|
FCRLB
|
Fc receptor-like B |
| chr19_+_19639704 | 0.04 |
ENST00000514277.4
|
YJEFN3
|
YjeF N-terminal domain containing 3 |
| chr14_-_75735986 | 0.04 |
ENST00000553510.1
|
RP11-293M10.1
|
Uncharacterized protein |
| chr5_-_13944652 | 0.04 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chr15_+_74422585 | 0.04 |
ENST00000561740.1
ENST00000435464.1 ENST00000453268.2 |
ISLR2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr11_+_44117260 | 0.04 |
ENST00000358681.4
|
EXT2
|
exostosin glycosyltransferase 2 |
| chr4_-_152149033 | 0.03 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr22_-_42739533 | 0.03 |
ENST00000515426.1
|
TCF20
|
transcription factor 20 (AR1) |
| chr5_+_140501581 | 0.03 |
ENST00000194152.1
|
PCDHB4
|
protocadherin beta 4 |
| chr7_-_115670804 | 0.03 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr12_+_49740700 | 0.03 |
ENST00000549441.2
ENST00000395069.3 |
DNAJC22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr10_-_74283694 | 0.03 |
ENST00000398763.4
ENST00000418483.2 ENST00000489666.2 |
MICU1
|
mitochondrial calcium uptake 1 |
| chr1_+_8378140 | 0.03 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr9_-_95298314 | 0.03 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr10_-_126716459 | 0.03 |
ENST00000309035.6
|
CTBP2
|
C-terminal binding protein 2 |
| chr10_-_128975273 | 0.03 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr1_+_47603109 | 0.03 |
ENST00000371890.3
ENST00000294337.3 ENST00000371891.3 |
CYP4A22
|
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr9_+_67977438 | 0.03 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU4F1_POU4F3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.3 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:1904457 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.2 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.3 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.0 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.0 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.6 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.0 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.1 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.2 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.6 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |