Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
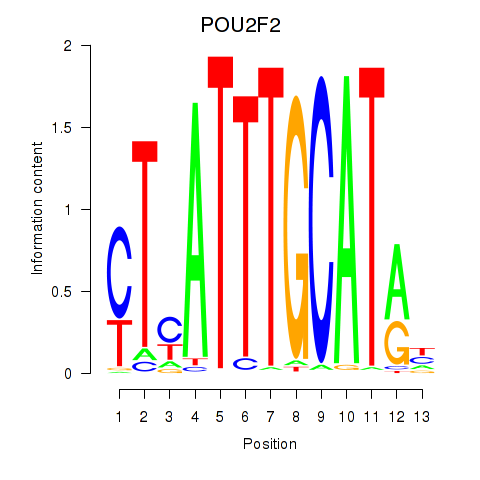
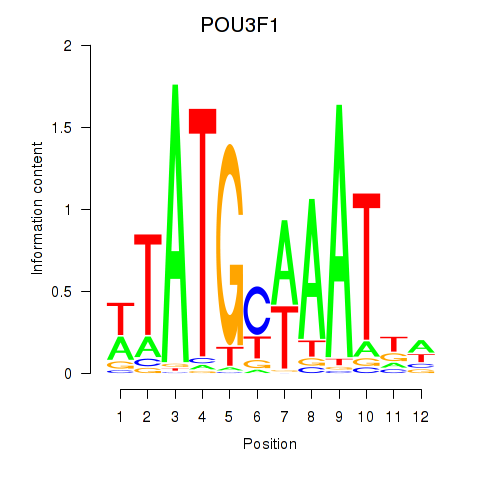
Results for POU2F2_POU3F1
Z-value: 1.39
Transcription factors associated with POU2F2_POU3F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F2
|
ENSG00000028277.16 | POU class 2 homeobox 2 |
|
POU3F1
|
ENSG00000185668.5 | POU class 3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F2 | hg19_v2_chr19_-_42636543_42636607 | -0.38 | 4.6e-01 | Click! |
Activity profile of POU2F2_POU3F1 motif
Sorted Z-values of POU2F2_POU3F1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_228645796 | 3.49 |
ENST00000369160.2
|
HIST3H2BB
|
histone cluster 3, H2bb |
| chr6_-_26124138 | 1.85 |
ENST00000314332.5
ENST00000396984.1 |
HIST1H2BC
|
histone cluster 1, H2bc |
| chr17_+_19091325 | 1.84 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr1_+_149858461 | 1.15 |
ENST00000331380.2
|
HIST2H2AC
|
histone cluster 2, H2ac |
| chr7_-_27153454 | 1.12 |
ENST00000522456.1
|
HOXA3
|
homeobox A3 |
| chr1_-_149783914 | 1.11 |
ENST00000369167.1
ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF
|
histone cluster 2, H2bf |
| chr6_-_26043885 | 1.02 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr17_-_7832753 | 1.01 |
ENST00000303790.2
|
KCNAB3
|
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr6_-_27775694 | 0.96 |
ENST00000377401.2
|
HIST1H2BL
|
histone cluster 1, H2bl |
| chr6_+_26217159 | 0.96 |
ENST00000303910.2
|
HIST1H2AE
|
histone cluster 1, H2ae |
| chr6_+_31588478 | 0.93 |
ENST00000376007.4
ENST00000376033.2 |
PRRC2A
|
proline-rich coiled-coil 2A |
| chr17_-_19015945 | 0.90 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr2_+_47922958 | 0.90 |
ENST00000606499.1
|
MSH6
|
mutS homolog 6 |
| chr9_-_35658007 | 0.84 |
ENST00000602361.1
|
RMRP
|
RNA component of mitochondrial RNA processing endoribonuclease |
| chr5_+_2752216 | 0.72 |
ENST00000457752.2
|
C5orf38
|
chromosome 5 open reading frame 38 |
| chr19_+_3572758 | 0.71 |
ENST00000416526.1
|
HMG20B
|
high mobility group 20B |
| chr16_-_73082274 | 0.71 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr6_-_27100529 | 0.65 |
ENST00000607124.1
ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ
|
histone cluster 1, H2bj |
| chr19_-_44124019 | 0.65 |
ENST00000300811.3
|
ZNF428
|
zinc finger protein 428 |
| chrX_-_55020511 | 0.63 |
ENST00000375006.3
ENST00000374992.2 |
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr19_-_50143452 | 0.62 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr12_-_58212487 | 0.60 |
ENST00000549994.1
|
AVIL
|
advillin |
| chr6_-_53013620 | 0.58 |
ENST00000259803.7
|
GCM1
|
glial cells missing homolog 1 (Drosophila) |
| chr6_-_138820624 | 0.58 |
ENST00000343505.5
|
NHSL1
|
NHS-like 1 |
| chr6_+_27861190 | 0.57 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr8_-_42396185 | 0.55 |
ENST00000518717.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr16_-_30107491 | 0.53 |
ENST00000566134.1
ENST00000565110.1 ENST00000398841.1 ENST00000398838.4 |
YPEL3
|
yippee-like 3 (Drosophila) |
| chr19_+_17516624 | 0.50 |
ENST00000596322.1
ENST00000600008.1 ENST00000601885.1 |
CTD-2521M24.9
|
CTD-2521M24.9 |
| chr14_-_68283291 | 0.50 |
ENST00000555452.1
ENST00000347230.4 |
ZFYVE26
|
zinc finger, FYVE domain containing 26 |
| chr19_-_4791219 | 0.46 |
ENST00000598782.1
|
AC005523.3
|
AC005523.3 |
| chr19_-_44123734 | 0.46 |
ENST00000598676.1
|
ZNF428
|
zinc finger protein 428 |
| chr14_-_106994333 | 0.45 |
ENST00000390624.2
|
IGHV3-48
|
immunoglobulin heavy variable 3-48 |
| chr19_+_56111680 | 0.45 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr16_-_21314360 | 0.44 |
ENST00000219599.3
ENST00000576703.1 |
CRYM
|
crystallin, mu |
| chr1_+_149822620 | 0.44 |
ENST00000369159.2
|
HIST2H2AA4
|
histone cluster 2, H2aa4 |
| chr19_+_19144666 | 0.43 |
ENST00000535288.1
ENST00000538663.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr19_-_45982076 | 0.43 |
ENST00000423698.2
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr19_+_19144384 | 0.43 |
ENST00000392335.2
ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6
|
armadillo repeat containing 6 |
| chr20_+_33292068 | 0.43 |
ENST00000374810.3
ENST00000374809.2 ENST00000451665.1 |
TP53INP2
|
tumor protein p53 inducible nuclear protein 2 |
| chr16_-_11730213 | 0.42 |
ENST00000576334.1
ENST00000574848.1 |
LITAF
|
lipopolysaccharide-induced TNF factor |
| chr16_-_88752889 | 0.41 |
ENST00000332281.5
|
SNAI3
|
snail family zinc finger 3 |
| chr12_+_7053228 | 0.41 |
ENST00000540506.2
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr22_-_43010859 | 0.39 |
ENST00000339677.6
|
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr19_-_46088068 | 0.38 |
ENST00000263275.4
ENST00000323060.3 |
OPA3
|
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr3_-_87039662 | 0.38 |
ENST00000494229.1
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr2_+_90192768 | 0.38 |
ENST00000390275.2
|
IGKV1D-13
|
immunoglobulin kappa variable 1D-13 |
| chr10_+_24528108 | 0.37 |
ENST00000438429.1
|
KIAA1217
|
KIAA1217 |
| chr12_-_51611477 | 0.36 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr7_+_26438187 | 0.35 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr12_-_120189900 | 0.35 |
ENST00000546026.1
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21) |
| chr5_+_158527630 | 0.35 |
ENST00000523301.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr16_-_74808710 | 0.35 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
| chr18_-_74839891 | 0.35 |
ENST00000581878.1
|
MBP
|
myelin basic protein |
| chr7_+_18535346 | 0.34 |
ENST00000405010.3
ENST00000406451.4 ENST00000428307.2 |
HDAC9
|
histone deacetylase 9 |
| chr17_+_76037081 | 0.34 |
ENST00000588549.1
|
TNRC6C
|
trinucleotide repeat containing 6C |
| chr12_+_7052974 | 0.34 |
ENST00000544681.1
ENST00000537087.1 |
C12orf57
|
chromosome 12 open reading frame 57 |
| chr11_-_64085533 | 0.33 |
ENST00000544844.1
|
TRMT112
|
tRNA methyltransferase 11-2 homolog (S. cerevisiae) |
| chr19_-_40791302 | 0.33 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chrX_-_106960285 | 0.33 |
ENST00000503515.1
ENST00000372397.2 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr14_-_51297360 | 0.33 |
ENST00000496749.1
|
NIN
|
ninein (GSK3B interacting protein) |
| chr22_-_43010928 | 0.32 |
ENST00000348657.2
ENST00000252115.5 |
POLDIP3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr22_+_38093005 | 0.32 |
ENST00000406386.3
|
TRIOBP
|
TRIO and F-actin binding protein |
| chr22_+_32750872 | 0.32 |
ENST00000397468.1
|
RFPL3
|
ret finger protein-like 3 |
| chr20_-_44298878 | 0.31 |
ENST00000324384.3
ENST00000356562.2 |
WFDC11
|
WAP four-disulfide core domain 11 |
| chr6_+_26273144 | 0.31 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr6_+_27782788 | 0.31 |
ENST00000359465.4
|
HIST1H2BM
|
histone cluster 1, H2bm |
| chr1_-_149859466 | 0.30 |
ENST00000331128.3
|
HIST2H2AB
|
histone cluster 2, H2ab |
| chr19_+_41305330 | 0.30 |
ENST00000593972.1
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr6_+_26183958 | 0.30 |
ENST00000356530.3
|
HIST1H2BE
|
histone cluster 1, H2be |
| chr3_+_187957646 | 0.30 |
ENST00000457242.1
|
LPP
|
LIM domain containing preferred translocation partner in lipoma |
| chr6_-_10415218 | 0.30 |
ENST00000466073.1
ENST00000498450.1 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr15_+_67458357 | 0.30 |
ENST00000537194.2
|
SMAD3
|
SMAD family member 3 |
| chr1_+_32538492 | 0.30 |
ENST00000336294.5
|
TMEM39B
|
transmembrane protein 39B |
| chr12_+_14369524 | 0.30 |
ENST00000538329.1
|
RP11-134N1.2
|
RP11-134N1.2 |
| chr5_+_158527485 | 0.29 |
ENST00000517335.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr22_+_45680822 | 0.29 |
ENST00000216211.4
ENST00000396082.2 |
UPK3A
|
uroplakin 3A |
| chr19_-_19144243 | 0.29 |
ENST00000594445.1
ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2
|
SURP and G patch domain containing 2 |
| chr12_+_7053172 | 0.29 |
ENST00000229281.5
|
C12orf57
|
chromosome 12 open reading frame 57 |
| chr21_+_38792602 | 0.29 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr9_+_92219919 | 0.28 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr4_-_156297919 | 0.28 |
ENST00000450097.1
|
MAP9
|
microtubule-associated protein 9 |
| chr1_+_201979645 | 0.28 |
ENST00000367284.5
ENST00000367283.3 |
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr19_+_41305085 | 0.28 |
ENST00000303961.4
|
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr1_+_32538520 | 0.28 |
ENST00000438825.1
ENST00000456834.2 ENST00000373634.4 ENST00000427288.1 |
TMEM39B
|
transmembrane protein 39B |
| chr2_+_66918558 | 0.27 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr11_+_64085560 | 0.27 |
ENST00000265462.4
ENST00000352435.4 ENST00000347941.4 |
PRDX5
|
peroxiredoxin 5 |
| chr14_+_93389425 | 0.27 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr5_+_158654712 | 0.27 |
ENST00000520323.1
|
CTB-11I22.2
|
CTB-11I22.2 |
| chr4_+_184826418 | 0.27 |
ENST00000308497.4
ENST00000438269.1 |
STOX2
|
storkhead box 2 |
| chr19_+_42788172 | 0.26 |
ENST00000160740.3
|
CIC
|
capicua transcriptional repressor |
| chr19_-_40791160 | 0.26 |
ENST00000358335.5
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr16_+_2198604 | 0.26 |
ENST00000210187.6
|
RAB26
|
RAB26, member RAS oncogene family |
| chr10_-_29811456 | 0.26 |
ENST00000535393.1
|
SVIL
|
supervillin |
| chr17_+_26638667 | 0.26 |
ENST00000600595.1
|
AC061975.10
|
Uncharacterized protein |
| chr11_+_74699942 | 0.25 |
ENST00000526068.1
ENST00000532963.1 ENST00000531619.1 ENST00000534628.1 ENST00000545272.1 |
NEU3
|
sialidase 3 (membrane sialidase) |
| chr10_-_10504285 | 0.25 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr1_-_149814478 | 0.24 |
ENST00000369161.3
|
HIST2H2AA3
|
histone cluster 2, H2aa3 |
| chr6_-_10415470 | 0.24 |
ENST00000379604.2
ENST00000379613.3 |
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr5_+_158527667 | 0.24 |
ENST00000499583.1
|
RP11-175K6.1
|
RP11-175K6.1 |
| chr1_+_17575584 | 0.24 |
ENST00000375460.3
|
PADI3
|
peptidyl arginine deiminase, type III |
| chr7_-_44122063 | 0.24 |
ENST00000335195.6
ENST00000395831.3 ENST00000414235.1 ENST00000452049.1 ENST00000242248.5 |
POLM
|
polymerase (DNA directed), mu |
| chr12_+_72080253 | 0.24 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr1_+_28099700 | 0.24 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr9_+_136325089 | 0.23 |
ENST00000291722.7
ENST00000316948.4 ENST00000540581.1 |
CACFD1
|
calcium channel flower domain containing 1 |
| chr7_-_100183742 | 0.23 |
ENST00000310300.6
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr3_-_87040259 | 0.23 |
ENST00000383698.3
|
VGLL3
|
vestigial like 3 (Drosophila) |
| chr6_-_128222103 | 0.23 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr5_+_60628074 | 0.23 |
ENST00000252744.5
|
ZSWIM6
|
zinc finger, SWIM-type containing 6 |
| chr4_-_187517928 | 0.23 |
ENST00000512772.1
|
FAT1
|
FAT atypical cadherin 1 |
| chr11_-_64084959 | 0.23 |
ENST00000535750.1
ENST00000535126.1 ENST00000539854.1 ENST00000308774.2 |
TRMT112
|
tRNA methyltransferase 11-2 homolog (S. cerevisiae) |
| chr2_+_220143989 | 0.22 |
ENST00000336576.5
|
DNAJB2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr17_+_38497640 | 0.22 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr12_+_107712173 | 0.22 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr16_+_3019309 | 0.22 |
ENST00000576565.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr7_+_18535321 | 0.22 |
ENST00000413380.1
ENST00000430454.1 |
HDAC9
|
histone deacetylase 9 |
| chrX_+_38660682 | 0.22 |
ENST00000457894.1
|
MID1IP1
|
MID1 interacting protein 1 |
| chr7_-_14029283 | 0.22 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr5_-_39462390 | 0.22 |
ENST00000511792.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_+_156052354 | 0.21 |
ENST00000368301.2
|
LMNA
|
lamin A/C |
| chr11_-_3663502 | 0.21 |
ENST00000359918.4
|
ART5
|
ADP-ribosyltransferase 5 |
| chr12_-_11091862 | 0.21 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr17_+_42925270 | 0.21 |
ENST00000253410.2
ENST00000587021.1 |
HIGD1B
|
HIG1 hypoxia inducible domain family, member 1B |
| chr6_+_43139037 | 0.20 |
ENST00000265354.4
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor) |
| chr17_+_72426891 | 0.20 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr1_+_24018269 | 0.20 |
ENST00000374550.3
|
RPL11
|
ribosomal protein L11 |
| chr7_+_100183927 | 0.20 |
ENST00000241071.6
ENST00000360609.2 |
FBXO24
|
F-box protein 24 |
| chr19_-_3971050 | 0.20 |
ENST00000545797.2
ENST00000596311.1 |
DAPK3
|
death-associated protein kinase 3 |
| chr2_-_239148599 | 0.20 |
ENST00000409182.1
ENST00000409002.3 ENST00000450098.1 ENST00000409356.1 ENST00000409160.3 ENST00000409574.1 ENST00000272937.5 |
HES6
|
hes family bHLH transcription factor 6 |
| chr17_-_46682321 | 0.20 |
ENST00000225648.3
ENST00000484302.2 |
HOXB6
|
homeobox B6 |
| chr15_-_89456593 | 0.19 |
ENST00000558029.1
ENST00000539437.1 ENST00000542878.1 ENST00000268151.7 ENST00000566497.1 |
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chr19_-_47291843 | 0.19 |
ENST00000542575.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr7_-_105332084 | 0.19 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr1_+_206579736 | 0.19 |
ENST00000439126.1
|
SRGAP2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr5_-_2751762 | 0.19 |
ENST00000302057.5
ENST00000382611.6 |
IRX2
|
iroquois homeobox 2 |
| chr2_-_863877 | 0.19 |
ENST00000415700.1
|
AC113607.1
|
long intergenic non-protein coding RNA 1115 |
| chr6_+_27833034 | 0.19 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr17_-_73127353 | 0.19 |
ENST00000580423.1
ENST00000578337.1 ENST00000582160.1 |
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr19_-_18392422 | 0.19 |
ENST00000252818.3
|
JUND
|
jun D proto-oncogene |
| chr19_-_893200 | 0.19 |
ENST00000269814.4
ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16
|
mediator complex subunit 16 |
| chr10_-_63995871 | 0.19 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr19_-_8373173 | 0.19 |
ENST00000537716.2
ENST00000301458.5 |
CD320
|
CD320 molecule |
| chr17_-_19651598 | 0.18 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr6_-_27860956 | 0.18 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chr17_+_73521763 | 0.18 |
ENST00000167462.5
ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr8_-_33370607 | 0.18 |
ENST00000360742.5
ENST00000523305.1 |
TTI2
|
TELO2 interacting protein 2 |
| chr2_+_54785485 | 0.18 |
ENST00000333896.5
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr12_-_122238464 | 0.18 |
ENST00000546227.1
|
RHOF
|
ras homolog family member F (in filopodia) |
| chr3_+_186435065 | 0.18 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr2_+_97426631 | 0.18 |
ENST00000377075.2
|
CNNM4
|
cyclin M4 |
| chr8_-_16859690 | 0.17 |
ENST00000180166.5
|
FGF20
|
fibroblast growth factor 20 |
| chr19_+_3572925 | 0.17 |
ENST00000333651.6
ENST00000417382.1 ENST00000453933.1 ENST00000262949.7 |
HMG20B
|
high mobility group 20B |
| chr11_-_62369291 | 0.17 |
ENST00000278823.2
|
MTA2
|
metastasis associated 1 family, member 2 |
| chr1_-_209792111 | 0.17 |
ENST00000455193.1
|
LAMB3
|
laminin, beta 3 |
| chrX_+_38660704 | 0.17 |
ENST00000378474.3
|
MID1IP1
|
MID1 interacting protein 1 |
| chr17_-_3794021 | 0.17 |
ENST00000381769.2
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha |
| chr18_-_52989217 | 0.17 |
ENST00000570287.2
|
TCF4
|
transcription factor 4 |
| chr3_-_165555200 | 0.17 |
ENST00000479451.1
ENST00000540653.1 ENST00000488954.1 ENST00000264381.3 |
BCHE
|
butyrylcholinesterase |
| chr11_-_75917569 | 0.17 |
ENST00000322563.3
|
WNT11
|
wingless-type MMTV integration site family, member 11 |
| chr20_+_32150140 | 0.17 |
ENST00000344201.3
ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr2_+_241938255 | 0.17 |
ENST00000401884.1
ENST00000405547.3 ENST00000310397.8 ENST00000342631.6 |
SNED1
|
sushi, nidogen and EGF-like domains 1 |
| chr12_+_130646999 | 0.16 |
ENST00000539839.1
ENST00000229030.4 |
FZD10
|
frizzled family receptor 10 |
| chr17_-_19651668 | 0.16 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr5_+_140593509 | 0.16 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr15_-_89456630 | 0.16 |
ENST00000268150.8
|
MFGE8
|
milk fat globule-EGF factor 8 protein |
| chr1_+_201979743 | 0.16 |
ENST00000446188.1
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
| chr19_+_50169081 | 0.15 |
ENST00000246784.3
|
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr20_+_816695 | 0.15 |
ENST00000246100.3
|
FAM110A
|
family with sequence similarity 110, member A |
| chr3_+_98250743 | 0.15 |
ENST00000284311.3
|
GPR15
|
G protein-coupled receptor 15 |
| chr19_+_17337027 | 0.15 |
ENST00000601529.1
ENST00000600232.1 |
OCEL1
|
occludin/ELL domain containing 1 |
| chr16_-_4164027 | 0.15 |
ENST00000572288.1
|
ADCY9
|
adenylate cyclase 9 |
| chr2_-_239197201 | 0.15 |
ENST00000254658.3
|
PER2
|
period circadian clock 2 |
| chr2_-_219906220 | 0.15 |
ENST00000458526.1
ENST00000409865.3 ENST00000410037.1 ENST00000457968.1 ENST00000436631.1 ENST00000341552.5 ENST00000441968.1 ENST00000295729.2 |
CCDC108
|
coiled-coil domain containing 108 |
| chr6_+_155538093 | 0.14 |
ENST00000462408.2
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr16_-_19897455 | 0.14 |
ENST00000568214.1
ENST00000569479.1 |
GPRC5B
|
G protein-coupled receptor, family C, group 5, member B |
| chr16_+_3019552 | 0.14 |
ENST00000572687.1
|
PAQR4
|
progestin and adipoQ receptor family member IV |
| chr19_-_1021113 | 0.14 |
ENST00000333175.5
ENST00000356663.3 |
TMEM259
|
transmembrane protein 259 |
| chr2_-_225811747 | 0.14 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr15_+_91478493 | 0.14 |
ENST00000418476.2
|
UNC45A
|
unc-45 homolog A (C. elegans) |
| chr6_+_27775899 | 0.14 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr10_+_123923205 | 0.13 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr17_-_73127826 | 0.13 |
ENST00000582170.1
ENST00000245552.2 |
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr12_+_51317788 | 0.13 |
ENST00000550502.1
|
METTL7A
|
methyltransferase like 7A |
| chr1_-_6052463 | 0.13 |
ENST00000378156.4
|
NPHP4
|
nephronophthisis 4 |
| chr2_-_239197238 | 0.13 |
ENST00000254657.3
|
PER2
|
period circadian clock 2 |
| chr5_-_131879205 | 0.13 |
ENST00000231454.1
|
IL5
|
interleukin 5 (colony-stimulating factor, eosinophil) |
| chr4_+_159727272 | 0.13 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chrX_+_154114635 | 0.13 |
ENST00000369446.2
|
F8A1
|
coagulation factor VIII-associated 1 |
| chr16_-_67867749 | 0.13 |
ENST00000566758.1
ENST00000445712.2 ENST00000219172.3 ENST00000564817.1 |
CENPT
|
centromere protein T |
| chr5_-_175965008 | 0.13 |
ENST00000537487.1
|
RNF44
|
ring finger protein 44 |
| chr22_-_22221900 | 0.13 |
ENST00000215832.6
ENST00000398822.3 |
MAPK1
|
mitogen-activated protein kinase 1 |
| chr19_-_41903161 | 0.13 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr1_+_207669495 | 0.13 |
ENST00000367052.1
ENST00000367051.1 ENST00000367053.1 |
CR1
|
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr2_+_3393453 | 0.13 |
ENST00000441983.1
|
TRAPPC12
|
trafficking protein particle complex 12 |
| chr2_-_61108449 | 0.13 |
ENST00000439412.1
ENST00000452343.1 |
AC010733.4
|
AC010733.4 |
| chr17_-_19651654 | 0.13 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chrX_-_154688276 | 0.13 |
ENST00000369445.2
|
F8A3
|
coagulation factor VIII-associated 3 |
| chr10_-_103880209 | 0.13 |
ENST00000425280.1
|
LDB1
|
LIM domain binding 1 |
| chr17_-_73127778 | 0.13 |
ENST00000578407.1
|
NT5C
|
5', 3'-nucleotidase, cytosolic |
| chr11_-_34379546 | 0.13 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr2_+_89952792 | 0.12 |
ENST00000390265.2
|
IGKV1D-33
|
immunoglobulin kappa variable 1D-33 |
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F2_POU3F1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 1.0 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.2 | 0.7 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 1.3 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.6 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 1.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.6 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.5 | GO:0021623 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.2 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.1 | 0.3 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.1 | 0.6 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.3 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.6 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.1 | 0.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.2 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.3 | GO:1904209 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.1 | 0.2 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.3 | GO:0030047 | actin modification(GO:0030047) |
| 0.1 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.1 | 0.2 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.2 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.2 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:0019249 | lactate biosynthetic process(GO:0019249) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.3 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.2 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.2 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.1 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.0 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.6 | GO:0090343 | positive regulation of cell aging(GO:0090343) positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.6 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 6.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.9 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.3 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.3 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.7 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.3 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) |
| 0.0 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:1902571 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.0 | GO:1905069 | allantois development(GO:1905069) |
| 0.0 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:2001191 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.2 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.4 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.0 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.3 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0090312 | positive regulation of protein deacetylation(GO:0090312) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.0 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.5 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.2 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 8.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.7 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.1 | 0.6 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.2 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 0.6 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 0.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.2 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 1.2 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0010736 | serum response element binding(GO:0010736) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.4 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.3 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.2 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.7 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0032396 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) inhibitory MHC class I receptor activity(GO:0032396) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 1.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 7.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |