Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
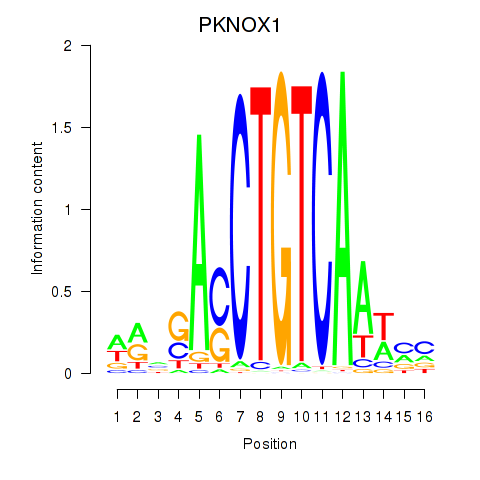
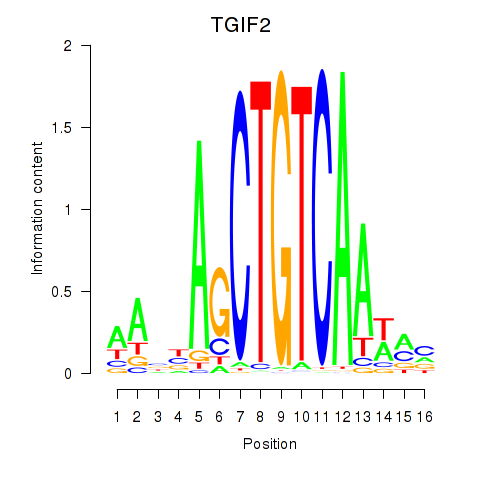
Results for PKNOX1_TGIF2
Z-value: 1.01
Transcription factors associated with PKNOX1_TGIF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX1
|
ENSG00000160199.10 | PBX/knotted 1 homeobox 1 |
|
TGIF2
|
ENSG00000118707.5 | TGFB induced factor homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF2 | hg19_v2_chr20_+_35202909_35203075 | 0.70 | 1.2e-01 | Click! |
| PKNOX1 | hg19_v2_chr21_+_44394620_44394737 | 0.07 | 8.9e-01 | Click! |
Activity profile of PKNOX1_TGIF2 motif
Sorted Z-values of PKNOX1_TGIF2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_39405939 | 0.68 |
ENST00000334109.2
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr7_+_23210760 | 0.67 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr9_-_134615326 | 0.59 |
ENST00000438647.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr1_+_207262578 | 0.58 |
ENST00000243611.5
ENST00000367076.3 |
C4BPB
|
complement component 4 binding protein, beta |
| chr11_+_6411670 | 0.57 |
ENST00000530395.1
ENST00000527275.1 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr1_+_207262881 | 0.57 |
ENST00000451804.2
|
C4BPB
|
complement component 4 binding protein, beta |
| chr7_-_74867509 | 0.56 |
ENST00000426327.3
|
GATSL2
|
GATS protein-like 2 |
| chr6_+_155334780 | 0.49 |
ENST00000538270.1
ENST00000535231.1 |
TIAM2
|
T-cell lymphoma invasion and metastasis 2 |
| chr3_+_111260980 | 0.49 |
ENST00000438817.2
|
CD96
|
CD96 molecule |
| chr22_+_17082732 | 0.45 |
ENST00000558085.2
ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1
|
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr7_+_70229899 | 0.43 |
ENST00000443672.1
|
AUTS2
|
autism susceptibility candidate 2 |
| chr11_+_6411636 | 0.41 |
ENST00000299397.3
ENST00000356761.2 ENST00000342245.4 |
SMPD1
|
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr4_-_46391367 | 0.41 |
ENST00000503806.1
ENST00000356504.1 ENST00000514090.1 ENST00000506961.1 |
GABRA2
|
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr17_-_77967433 | 0.39 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr10_-_4720301 | 0.35 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr19_+_36393367 | 0.35 |
ENST00000246551.4
|
HCST
|
hematopoietic cell signal transducer |
| chr13_+_24844857 | 0.33 |
ENST00000409126.1
ENST00000343003.6 |
SPATA13
|
spermatogenesis associated 13 |
| chr22_+_20905269 | 0.33 |
ENST00000457322.1
|
MED15
|
mediator complex subunit 15 |
| chr3_-_52719912 | 0.33 |
ENST00000420148.1
|
PBRM1
|
polybromo 1 |
| chr18_+_54318893 | 0.33 |
ENST00000593058.1
|
WDR7
|
WD repeat domain 7 |
| chr14_+_61995722 | 0.31 |
ENST00000556347.1
|
RP11-47I22.4
|
RP11-47I22.4 |
| chr3_-_52719810 | 0.31 |
ENST00000424867.1
ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1
|
polybromo 1 |
| chr2_-_192015697 | 0.31 |
ENST00000409995.1
|
STAT4
|
signal transducer and activator of transcription 4 |
| chr4_-_175443788 | 0.31 |
ENST00000541923.1
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr6_+_41021027 | 0.31 |
ENST00000244669.2
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr11_+_827553 | 0.31 |
ENST00000528542.2
ENST00000450448.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr1_+_154966058 | 0.31 |
ENST00000392487.1
|
LENEP
|
lens epithelial protein |
| chr5_-_94620239 | 0.29 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr16_-_79634595 | 0.29 |
ENST00000326043.4
ENST00000393350.1 |
MAF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr15_+_59908633 | 0.29 |
ENST00000559626.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr18_+_54318566 | 0.28 |
ENST00000589935.1
ENST00000357574.3 |
WDR7
|
WD repeat domain 7 |
| chr1_+_186649754 | 0.28 |
ENST00000608917.1
|
RP5-973M2.2
|
RP5-973M2.2 |
| chr17_-_65992544 | 0.27 |
ENST00000580729.1
|
RP11-855A2.5
|
RP11-855A2.5 |
| chr19_+_9296279 | 0.27 |
ENST00000344248.2
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2 |
| chr2_-_190446738 | 0.27 |
ENST00000427419.1
ENST00000455320.1 |
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr16_+_20817953 | 0.26 |
ENST00000568647.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr3_-_52488048 | 0.26 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr12_-_11036844 | 0.26 |
ENST00000428168.2
|
PRH1
|
proline-rich protein HaeIII subfamily 1 |
| chr4_-_105416039 | 0.26 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr7_-_99869799 | 0.26 |
ENST00000436886.2
|
GATS
|
GATS, stromal antigen 3 opposite strand |
| chr12_+_85430110 | 0.25 |
ENST00000393212.3
ENST00000393217.2 |
LRRIQ1
|
leucine-rich repeats and IQ motif containing 1 |
| chr1_+_203765437 | 0.25 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr17_-_29151686 | 0.24 |
ENST00000544695.1
|
CRLF3
|
cytokine receptor-like factor 3 |
| chr18_-_44775554 | 0.24 |
ENST00000425639.1
ENST00000400404.1 |
SKOR2
|
SKI family transcriptional corepressor 2 |
| chr2_+_29001711 | 0.24 |
ENST00000418910.1
|
PPP1CB
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr16_+_20817746 | 0.24 |
ENST00000568894.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr5_+_173763250 | 0.24 |
ENST00000515513.1
ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1
|
RP11-267A15.1 |
| chr5_+_125758813 | 0.24 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr17_-_72358001 | 0.23 |
ENST00000375366.3
|
BTBD17
|
BTB (POZ) domain containing 17 |
| chrY_-_21239221 | 0.23 |
ENST00000447937.1
ENST00000331787.2 |
TTTY14
|
testis-specific transcript, Y-linked 14 (non-protein coding) |
| chr1_-_179198806 | 0.22 |
ENST00000392043.3
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr22_+_20905422 | 0.22 |
ENST00000424287.1
ENST00000423862.1 |
MED15
|
mediator complex subunit 15 |
| chr11_+_120207787 | 0.21 |
ENST00000397843.2
ENST00000356641.3 |
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr1_+_104104379 | 0.21 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr6_+_111195973 | 0.21 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chr19_-_54604083 | 0.20 |
ENST00000391761.1
ENST00000356532.3 ENST00000359649.4 ENST00000358375.4 ENST00000391760.1 ENST00000351806.4 |
OSCAR
|
osteoclast associated, immunoglobulin-like receptor |
| chr1_-_145715565 | 0.20 |
ENST00000369288.2
ENST00000369290.1 ENST00000401557.3 |
CD160
|
CD160 molecule |
| chr12_+_69186125 | 0.20 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr12_-_31882108 | 0.20 |
ENST00000281471.6
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr1_-_214638146 | 0.20 |
ENST00000543945.1
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr16_-_30569801 | 0.20 |
ENST00000395091.2
|
ZNF764
|
zinc finger protein 764 |
| chr3_-_27764190 | 0.20 |
ENST00000537516.1
|
EOMES
|
eomesodermin |
| chr12_-_105478339 | 0.20 |
ENST00000424857.2
ENST00000258494.9 |
ALDH1L2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr5_+_125758865 | 0.20 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_125759140 | 0.20 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr16_-_30457048 | 0.20 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr17_-_34122596 | 0.19 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr17_-_8093471 | 0.19 |
ENST00000389017.4
|
C17orf59
|
chromosome 17 open reading frame 59 |
| chr1_+_24285599 | 0.19 |
ENST00000471915.1
|
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr10_-_99052382 | 0.19 |
ENST00000453547.2
ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1
ARHGAP19
|
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr22_+_39052632 | 0.19 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr9_-_139345270 | 0.18 |
ENST00000398335.1
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr1_-_179198702 | 0.18 |
ENST00000502732.1
|
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr5_-_176449586 | 0.18 |
ENST00000509236.1
|
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr15_-_31283798 | 0.17 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr2_-_74779744 | 0.17 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr20_-_36156264 | 0.17 |
ENST00000445723.1
ENST00000414080.1 |
BLCAP
|
bladder cancer associated protein |
| chr6_-_163148780 | 0.16 |
ENST00000366892.1
ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr15_-_76352069 | 0.16 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr3_-_113160334 | 0.16 |
ENST00000393845.2
ENST00000295868.2 |
WDR52
|
WD repeat domain 52 |
| chr4_+_146539415 | 0.16 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr3_-_111852061 | 0.16 |
ENST00000488580.1
ENST00000460387.2 ENST00000484193.1 ENST00000487901.1 |
GCSAM
|
germinal center-associated, signaling and motility |
| chr5_-_66942617 | 0.16 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr5_-_154230130 | 0.16 |
ENST00000519501.1
ENST00000518651.1 ENST00000517938.1 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chr9_-_99180597 | 0.15 |
ENST00000375256.4
|
ZNF367
|
zinc finger protein 367 |
| chr17_-_71223839 | 0.15 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr9_+_123906331 | 0.15 |
ENST00000431571.1
|
CNTRL
|
centriolin |
| chr15_-_37392086 | 0.15 |
ENST00000561208.1
|
MEIS2
|
Meis homeobox 2 |
| chr15_+_43477455 | 0.15 |
ENST00000300213.4
|
CCNDBP1
|
cyclin D-type binding-protein 1 |
| chr1_+_246729815 | 0.15 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr4_+_3076388 | 0.15 |
ENST00000355072.5
|
HTT
|
huntingtin |
| chr2_+_242127924 | 0.15 |
ENST00000402530.3
ENST00000274979.8 ENST00000402430.3 |
ANO7
|
anoctamin 7 |
| chr2_-_61765732 | 0.15 |
ENST00000443240.1
ENST00000436018.1 |
XPO1
|
exportin 1 (CRM1 homolog, yeast) |
| chr4_-_47983519 | 0.15 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr17_-_34195862 | 0.14 |
ENST00000592980.1
ENST00000587626.1 |
C17orf66
|
chromosome 17 open reading frame 66 |
| chr7_+_73442487 | 0.14 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr20_-_33872518 | 0.14 |
ENST00000374436.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr4_-_175443484 | 0.14 |
ENST00000514584.1
ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr6_-_31651817 | 0.14 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr3_+_185080908 | 0.14 |
ENST00000265026.3
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr6_-_28220002 | 0.14 |
ENST00000377294.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr11_-_67415048 | 0.14 |
ENST00000529256.1
|
ACY3
|
aspartoacylase (aminocyclase) 3 |
| chr3_+_38017264 | 0.14 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr13_-_46716969 | 0.14 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr19_-_58951496 | 0.14 |
ENST00000254166.3
|
ZNF132
|
zinc finger protein 132 |
| chr5_+_43603229 | 0.14 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr2_+_113321939 | 0.13 |
ENST00000458012.2
|
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr7_+_76139833 | 0.13 |
ENST00000257632.5
|
UPK3B
|
uroplakin 3B |
| chr3_-_142166846 | 0.13 |
ENST00000463916.1
ENST00000544157.1 |
XRN1
|
5'-3' exoribonuclease 1 |
| chr1_+_120049826 | 0.13 |
ENST00000369413.3
ENST00000235547.6 ENST00000528909.1 |
HSD3B1
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr13_+_24844819 | 0.13 |
ENST00000399949.2
|
SPATA13
|
spermatogenesis associated 13 |
| chr3_+_63897605 | 0.13 |
ENST00000487717.1
|
ATXN7
|
ataxin 7 |
| chr12_-_31882027 | 0.13 |
ENST00000541931.1
ENST00000535408.1 |
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr5_-_36301984 | 0.13 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr2_-_3504587 | 0.13 |
ENST00000415131.1
|
ADI1
|
acireductone dioxygenase 1 |
| chr2_+_217082311 | 0.13 |
ENST00000597904.1
|
RP11-566E18.3
|
RP11-566E18.3 |
| chr12_-_67197760 | 0.13 |
ENST00000539540.1
ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr7_-_53879623 | 0.12 |
ENST00000380970.2
|
GS1-179L18.1
|
GS1-179L18.1 |
| chr9_+_125027127 | 0.12 |
ENST00000441707.1
ENST00000373723.5 ENST00000373729.1 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr21_+_30502806 | 0.12 |
ENST00000399928.1
ENST00000399926.1 |
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr7_+_76139741 | 0.12 |
ENST00000334348.3
ENST00000419923.2 ENST00000448265.3 ENST00000443097.2 |
UPK3B
|
uroplakin 3B |
| chr3_-_52569023 | 0.12 |
ENST00000307076.4
|
NT5DC2
|
5'-nucleotidase domain containing 2 |
| chr21_+_30503282 | 0.12 |
ENST00000399925.1
|
MAP3K7CL
|
MAP3K7 C-terminal like |
| chr17_+_76374714 | 0.12 |
ENST00000262764.6
ENST00000589689.1 ENST00000329897.7 ENST00000592043.1 ENST00000587356.1 |
PGS1
|
phosphatidylglycerophosphate synthase 1 |
| chr8_+_11666649 | 0.12 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_-_225811747 | 0.12 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr1_-_2323140 | 0.12 |
ENST00000378531.3
ENST00000378529.3 |
MORN1
|
MORN repeat containing 1 |
| chr5_-_58882219 | 0.12 |
ENST00000505453.1
ENST00000360047.5 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr11_+_8932828 | 0.12 |
ENST00000530281.1
ENST00000396648.2 ENST00000534147.1 ENST00000529942.1 |
AKIP1
|
A kinase (PRKA) interacting protein 1 |
| chr7_+_29234375 | 0.11 |
ENST00000409350.1
ENST00000495789.2 ENST00000539389.1 |
CHN2
|
chimerin 2 |
| chr16_+_27413483 | 0.11 |
ENST00000337929.3
ENST00000564089.1 |
IL21R
|
interleukin 21 receptor |
| chr1_+_207262627 | 0.11 |
ENST00000391923.1
|
C4BPB
|
complement component 4 binding protein, beta |
| chr1_-_247171347 | 0.11 |
ENST00000339986.7
ENST00000487338.2 |
ZNF695
|
zinc finger protein 695 |
| chr16_-_89556942 | 0.11 |
ENST00000301030.4
|
ANKRD11
|
ankyrin repeat domain 11 |
| chr2_+_10560147 | 0.11 |
ENST00000422133.1
|
HPCAL1
|
hippocalcin-like 1 |
| chrX_-_148713440 | 0.11 |
ENST00000536359.1
ENST00000316916.8 |
TMEM185A
|
transmembrane protein 185A |
| chr6_-_86099898 | 0.11 |
ENST00000455071.1
|
RP11-30P6.6
|
RP11-30P6.6 |
| chr3_-_33481835 | 0.11 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr11_-_107729504 | 0.11 |
ENST00000265836.7
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr14_-_21994525 | 0.11 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr2_-_179914760 | 0.10 |
ENST00000420890.2
ENST00000409284.1 ENST00000443758.1 ENST00000446116.1 |
CCDC141
|
coiled-coil domain containing 141 |
| chr3_+_183899770 | 0.10 |
ENST00000442686.1
|
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr9_-_123639600 | 0.10 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr12_+_95611536 | 0.10 |
ENST00000549002.1
|
VEZT
|
vezatin, adherens junctions transmembrane protein |
| chr19_+_49375649 | 0.10 |
ENST00000200453.5
|
PPP1R15A
|
protein phosphatase 1, regulatory subunit 15A |
| chr3_+_9691117 | 0.10 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr17_+_13972807 | 0.10 |
ENST00000429152.2
ENST00000261643.3 ENST00000536205.1 ENST00000537334.1 |
COX10
|
cytochrome c oxidase assembly homolog 10 (yeast) |
| chr1_-_179112173 | 0.10 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr5_+_118407053 | 0.10 |
ENST00000311085.8
ENST00000539542.1 |
DMXL1
|
Dmx-like 1 |
| chr2_-_242242133 | 0.10 |
ENST00000430918.1
|
HDLBP
|
high density lipoprotein binding protein |
| chr15_-_37391614 | 0.10 |
ENST00000219869.9
|
MEIS2
|
Meis homeobox 2 |
| chr15_-_78112553 | 0.10 |
ENST00000562933.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr1_+_110577229 | 0.10 |
ENST00000369795.3
ENST00000369794.2 |
STRIP1
|
striatin interacting protein 1 |
| chr2_-_152589670 | 0.10 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr19_+_49496705 | 0.10 |
ENST00000595090.1
|
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr1_-_54872059 | 0.09 |
ENST00000371320.3
|
SSBP3
|
single stranded DNA binding protein 3 |
| chr4_+_128886584 | 0.09 |
ENST00000513371.1
|
C4orf29
|
chromosome 4 open reading frame 29 |
| chr16_+_50776021 | 0.09 |
ENST00000566679.2
ENST00000564634.1 ENST00000398568.2 |
CYLD
|
cylindromatosis (turban tumor syndrome) |
| chr17_+_38296576 | 0.09 |
ENST00000264645.7
|
CASC3
|
cancer susceptibility candidate 3 |
| chr1_-_179112189 | 0.09 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr1_-_32384693 | 0.09 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr16_-_2260834 | 0.09 |
ENST00000562360.1
ENST00000566018.1 |
BRICD5
|
BRICHOS domain containing 5 |
| chrX_+_135730373 | 0.09 |
ENST00000370628.2
|
CD40LG
|
CD40 ligand |
| chr16_+_20817926 | 0.09 |
ENST00000565340.1
|
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr6_+_163148973 | 0.09 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr8_-_67090825 | 0.09 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
| chr1_-_247335269 | 0.09 |
ENST00000543802.2
ENST00000491356.1 ENST00000472531.1 ENST00000340684.6 |
ZNF124
|
zinc finger protein 124 |
| chr3_+_57875711 | 0.09 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr6_-_84418432 | 0.09 |
ENST00000519825.1
ENST00000523484.2 |
SNAP91
|
synaptosomal-associated protein, 91kDa |
| chr1_-_45672221 | 0.09 |
ENST00000359600.5
|
ZSWIM5
|
zinc finger, SWIM-type containing 5 |
| chr17_+_78518617 | 0.09 |
ENST00000537330.1
ENST00000570891.1 |
RPTOR
|
regulatory associated protein of MTOR, complex 1 |
| chr9_+_123884038 | 0.09 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr22_+_35776354 | 0.09 |
ENST00000412893.1
|
HMOX1
|
heme oxygenase (decycling) 1 |
| chr20_-_33872548 | 0.09 |
ENST00000374443.3
|
EIF6
|
eukaryotic translation initiation factor 6 |
| chr12_+_57522692 | 0.09 |
ENST00000554174.1
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr12_-_71003568 | 0.09 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr4_-_175443943 | 0.09 |
ENST00000296522.6
|
HPGD
|
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr5_-_81046841 | 0.09 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr10_-_16859442 | 0.09 |
ENST00000602389.1
ENST00000345264.5 |
RSU1
|
Ras suppressor protein 1 |
| chr1_+_171217622 | 0.08 |
ENST00000433267.1
ENST00000367750.3 |
FMO1
|
flavin containing monooxygenase 1 |
| chr11_-_46867780 | 0.08 |
ENST00000529230.1
ENST00000415402.1 ENST00000312055.5 |
CKAP5
|
cytoskeleton associated protein 5 |
| chr14_-_21492251 | 0.08 |
ENST00000554398.1
|
NDRG2
|
NDRG family member 2 |
| chrX_+_135730297 | 0.08 |
ENST00000370629.2
|
CD40LG
|
CD40 ligand |
| chr16_+_4896659 | 0.08 |
ENST00000592120.1
|
UBN1
|
ubinuclein 1 |
| chr12_+_69201923 | 0.08 |
ENST00000462284.1
ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr17_-_35969409 | 0.08 |
ENST00000394378.2
ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG
|
synergin, gamma |
| chr6_-_24489842 | 0.08 |
ENST00000230036.1
|
GPLD1
|
glycosylphosphatidylinositol specific phospholipase D1 |
| chr10_-_79398127 | 0.08 |
ENST00000372443.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr21_+_38792602 | 0.08 |
ENST00000398960.2
ENST00000398956.2 |
DYRK1A
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr19_-_45663408 | 0.08 |
ENST00000317951.4
|
NKPD1
|
NTPase, KAP family P-loop domain containing 1 |
| chr5_+_40841276 | 0.08 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr1_+_149824160 | 0.08 |
ENST00000403683.1
|
HIST2H3A
|
histone cluster 2, H3a |
| chr2_-_175260368 | 0.08 |
ENST00000342016.3
ENST00000362053.5 |
CIR1
|
corepressor interacting with RBPJ, 1 |
| chr22_+_50946645 | 0.08 |
ENST00000420993.2
ENST00000395698.3 ENST00000395701.3 ENST00000523045.1 ENST00000299821.11 |
NCAPH2
|
non-SMC condensin II complex, subunit H2 |
| chr7_-_156685841 | 0.08 |
ENST00000354505.4
ENST00000540390.1 |
LMBR1
|
limb development membrane protein 1 |
| chr16_+_20818020 | 0.08 |
ENST00000564274.1
ENST00000563068.1 |
AC004381.6
|
Putative RNA exonuclease NEF-sp |
| chr8_-_8318847 | 0.08 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr6_-_163148700 | 0.07 |
ENST00000366894.1
ENST00000338468.3 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr5_-_55008101 | 0.07 |
ENST00000506624.1
ENST00000513275.1 ENST00000513993.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr19_-_13044494 | 0.07 |
ENST00000593021.1
ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA
|
phenylalanyl-tRNA synthetase, alpha subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX1_TGIF2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.2 | 1.4 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 1.0 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.1 | 0.4 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.1 | 0.3 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.3 | GO:1903414 | spleen trabecula formation(GO:0060345) iron cation export(GO:1903414) ferrous iron export(GO:1903988) |
| 0.1 | 0.2 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.5 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 0.2 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 0.1 | 0.2 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.2 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.3 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:2000742 | anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) cellular response to cocaine(GO:0071314) positive regulation of fear response(GO:1903367) regulation of corticosterone secretion(GO:2000852) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.2 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.6 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.2 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0016259 | seryl-tRNA aminoacylation(GO:0006434) selenocysteine metabolic process(GO:0016259) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0010982 | negative regulation of triglyceride catabolic process(GO:0010897) regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:0036215 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0050960 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.0 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.0 | GO:0070175 | positive regulation of enamel mineralization(GO:0070175) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.8 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.3 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.3 | GO:0097689 | iron channel activity(GO:0097689) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.1 | 0.2 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.4 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |