Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
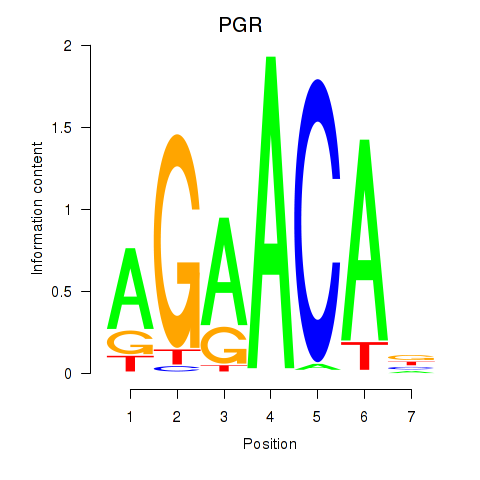
Results for PGR
Z-value: 0.67
Transcription factors associated with PGR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PGR
|
ENSG00000082175.10 | progesterone receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PGR | hg19_v2_chr11_-_100999775_100999801 | -0.23 | 6.6e-01 | Click! |
Activity profile of PGR motif
Sorted Z-values of PGR motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_615570 | 0.60 |
ENST00000525445.1
ENST00000348655.6 ENST00000397566.1 |
IRF7
|
interferon regulatory factor 7 |
| chr1_+_209859510 | 0.50 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr7_-_127672146 | 0.47 |
ENST00000476782.1
|
LRRC4
|
leucine rich repeat containing 4 |
| chr7_-_5821225 | 0.42 |
ENST00000416985.1
|
RNF216
|
ring finger protein 216 |
| chr14_+_24641062 | 0.40 |
ENST00000311457.3
ENST00000557806.1 ENST00000559919.1 |
REC8
|
REC8 meiotic recombination protein |
| chr6_+_76330355 | 0.34 |
ENST00000483859.2
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr2_+_38177575 | 0.34 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chrX_+_38420623 | 0.32 |
ENST00000378482.2
|
TSPAN7
|
tetraspanin 7 |
| chr17_+_39846114 | 0.32 |
ENST00000586699.1
|
EIF1
|
eukaryotic translation initiation factor 1 |
| chr16_-_3350614 | 0.32 |
ENST00000268674.2
|
TIGD7
|
tigger transposable element derived 7 |
| chr12_+_21654714 | 0.30 |
ENST00000542038.1
ENST00000540141.1 ENST00000229314.5 |
GOLT1B
|
golgi transport 1B |
| chr2_+_9778872 | 0.28 |
ENST00000478468.1
|
RP11-521D12.1
|
RP11-521D12.1 |
| chr6_-_150212029 | 0.28 |
ENST00000529948.1
ENST00000357183.4 ENST00000367363.3 |
RAET1E
|
retinoic acid early transcript 1E |
| chr3_-_149051194 | 0.27 |
ENST00000470080.1
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr15_-_29745003 | 0.27 |
ENST00000560082.1
|
FAM189A1
|
family with sequence similarity 189, member A1 |
| chr3_-_114035026 | 0.26 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr4_+_108815402 | 0.25 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr3_-_49722523 | 0.25 |
ENST00000448220.1
|
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr2_-_219925189 | 0.25 |
ENST00000295731.6
|
IHH
|
indian hedgehog |
| chr14_+_29299437 | 0.24 |
ENST00000550827.1
ENST00000548087.1 ENST00000551588.1 ENST00000550122.1 |
CTD-2384A14.1
|
CTD-2384A14.1 |
| chr12_+_57849048 | 0.24 |
ENST00000266646.2
|
INHBE
|
inhibin, beta E |
| chr12_-_7244469 | 0.24 |
ENST00000538050.1
ENST00000536053.2 |
C1R
|
complement component 1, r subcomponent |
| chr10_-_94301107 | 0.23 |
ENST00000436178.1
|
IDE
|
insulin-degrading enzyme |
| chr11_+_35222629 | 0.23 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr5_+_148442844 | 0.21 |
ENST00000515519.1
|
CTC-529P8.1
|
CTC-529P8.1 |
| chr6_+_27107053 | 0.21 |
ENST00000354348.2
|
HIST1H4I
|
histone cluster 1, H4i |
| chr4_-_89442940 | 0.21 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr2_-_74776586 | 0.21 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr9_+_127615733 | 0.21 |
ENST00000373574.1
|
WDR38
|
WD repeat domain 38 |
| chr4_+_79567362 | 0.20 |
ENST00000512322.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr2_-_179315786 | 0.20 |
ENST00000457633.1
ENST00000438687.3 ENST00000325748.4 |
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr4_+_86525299 | 0.19 |
ENST00000512201.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_-_76203454 | 0.19 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr4_+_39699664 | 0.19 |
ENST00000261427.5
ENST00000510934.1 ENST00000295963.6 |
UBE2K
|
ubiquitin-conjugating enzyme E2K |
| chr7_+_22766766 | 0.19 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr13_-_52026730 | 0.19 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr13_-_52027134 | 0.18 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr4_-_146019693 | 0.18 |
ENST00000514390.1
|
ANAPC10
|
anaphase promoting complex subunit 10 |
| chr18_+_61445007 | 0.18 |
ENST00000447428.1
ENST00000546027.1 |
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr12_-_125401885 | 0.18 |
ENST00000542416.1
|
UBC
|
ubiquitin C |
| chr3_-_149510553 | 0.18 |
ENST00000462519.2
ENST00000446160.1 ENST00000383050.3 |
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1 |
| chr7_-_14026063 | 0.18 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr10_+_73156664 | 0.17 |
ENST00000398809.4
ENST00000398842.3 ENST00000461841.3 ENST00000299366.7 |
CDH23
|
cadherin-related 23 |
| chr12_+_28410128 | 0.17 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr2_-_99797390 | 0.17 |
ENST00000422537.2
|
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr8_+_27184320 | 0.17 |
ENST00000522517.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr2_+_60983361 | 0.17 |
ENST00000238714.3
|
PAPOLG
|
poly(A) polymerase gamma |
| chr8_-_82644562 | 0.17 |
ENST00000520604.1
ENST00000521742.1 ENST00000520635.1 |
ZFAND1
|
zinc finger, AN1-type domain 1 |
| chr1_+_172422026 | 0.17 |
ENST00000367725.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr17_-_70053866 | 0.16 |
ENST00000540802.1
|
RP11-84E24.2
|
RP11-84E24.2 |
| chr2_+_232135245 | 0.16 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr2_+_234601512 | 0.16 |
ENST00000305139.6
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr13_+_78315528 | 0.16 |
ENST00000496045.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr5_-_95018660 | 0.16 |
ENST00000395899.3
ENST00000274432.8 |
SPATA9
|
spermatogenesis associated 9 |
| chr14_-_35182994 | 0.16 |
ENST00000341223.3
|
CFL2
|
cofilin 2 (muscle) |
| chr8_-_110346614 | 0.15 |
ENST00000239690.4
|
NUDCD1
|
NudC domain containing 1 |
| chr1_+_154975258 | 0.15 |
ENST00000417934.2
|
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr7_-_135433460 | 0.15 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr3_-_149051444 | 0.15 |
ENST00000296059.2
|
TM4SF18
|
transmembrane 4 L six family member 18 |
| chr17_+_39411636 | 0.15 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr6_-_113953705 | 0.15 |
ENST00000452675.1
|
RP11-367G18.1
|
RP11-367G18.1 |
| chr4_+_79567057 | 0.15 |
ENST00000503259.1
ENST00000507802.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr11_-_113577014 | 0.14 |
ENST00000544634.1
ENST00000539732.1 ENST00000538770.1 ENST00000536856.1 ENST00000544476.1 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chrX_-_114953669 | 0.14 |
ENST00000449327.1
|
RP1-241P17.4
|
Uncharacterized protein |
| chr6_-_32157947 | 0.14 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr5_-_79946820 | 0.14 |
ENST00000604882.1
|
MTRNR2L2
|
MT-RNR2-like 2 |
| chr4_+_79567314 | 0.14 |
ENST00000503539.1
ENST00000504675.1 |
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr3_+_137906154 | 0.14 |
ENST00000466749.1
ENST00000358441.2 ENST00000489213.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr16_+_57680043 | 0.14 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chrX_-_133792480 | 0.14 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr1_+_42922173 | 0.14 |
ENST00000455780.1
ENST00000372560.3 ENST00000372561.3 ENST00000372556.3 |
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr15_-_49255632 | 0.14 |
ENST00000332408.4
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr11_-_10920838 | 0.14 |
ENST00000503469.2
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr1_+_212475148 | 0.13 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr1_+_78470530 | 0.13 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr7_-_75241096 | 0.13 |
ENST00000420909.1
|
HIP1
|
huntingtin interacting protein 1 |
| chr2_-_183387064 | 0.13 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr10_-_97200772 | 0.13 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr2_-_99797473 | 0.13 |
ENST00000409107.1
ENST00000289359.2 |
MITD1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr10_+_80008505 | 0.12 |
ENST00000434974.1
ENST00000423770.1 ENST00000432742.1 |
LINC00856
|
long intergenic non-protein coding RNA 856 |
| chr2_+_234602305 | 0.12 |
ENST00000406651.1
|
UGT1A6
|
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr5_+_64064748 | 0.12 |
ENST00000381070.3
ENST00000508024.1 |
CWC27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr2_-_179315490 | 0.12 |
ENST00000487082.1
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr6_+_33551515 | 0.12 |
ENST00000374458.1
|
GGNBP1
|
gametogenetin binding protein 1 (pseudogene) |
| chr4_-_23735183 | 0.12 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr11_-_10530723 | 0.12 |
ENST00000536684.1
|
MTRNR2L8
|
MT-RNR2-like 8 |
| chr11_+_111169565 | 0.12 |
ENST00000528846.1
|
COLCA2
|
colorectal cancer associated 2 |
| chr4_-_71532339 | 0.12 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr2_-_216300784 | 0.12 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr3_+_149531607 | 0.12 |
ENST00000468648.1
ENST00000459632.1 |
RNF13
|
ring finger protein 13 |
| chr3_+_149530836 | 0.11 |
ENST00000466478.1
ENST00000491086.1 ENST00000467977.1 |
RNF13
|
ring finger protein 13 |
| chr4_-_87028478 | 0.11 |
ENST00000515400.1
ENST00000395157.3 |
MAPK10
|
mitogen-activated protein kinase 10 |
| chr1_+_150229554 | 0.11 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr19_+_13135731 | 0.11 |
ENST00000587260.1
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr11_+_134225909 | 0.11 |
ENST00000533324.1
|
GLB1L2
|
galactosidase, beta 1-like 2 |
| chr6_-_76203345 | 0.11 |
ENST00000393004.2
|
FILIP1
|
filamin A interacting protein 1 |
| chr7_+_30960915 | 0.11 |
ENST00000441328.2
ENST00000409899.1 ENST00000409611.1 |
AQP1
|
aquaporin 1 (Colton blood group) |
| chr6_-_10413112 | 0.11 |
ENST00000465858.1
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr1_+_1846519 | 0.11 |
ENST00000378604.3
|
CALML6
|
calmodulin-like 6 |
| chr5_+_102201430 | 0.11 |
ENST00000438793.3
ENST00000346918.2 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr9_-_15510989 | 0.11 |
ENST00000380715.1
ENST00000380716.4 ENST00000380738.4 ENST00000380733.4 |
PSIP1
|
PC4 and SFRS1 interacting protein 1 |
| chr4_+_128802016 | 0.11 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr7_+_99202003 | 0.11 |
ENST00000609449.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr6_-_128222103 | 0.11 |
ENST00000434358.1
ENST00000543064.1 ENST00000368248.2 |
THEMIS
|
thymocyte selection associated |
| chr4_+_79567233 | 0.11 |
ENST00000514130.1
|
RP11-792D21.2
|
long intergenic non-protein coding RNA 1094 |
| chr13_+_102104980 | 0.11 |
ENST00000545560.2
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr7_-_14026123 | 0.11 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
| chr4_-_71705027 | 0.11 |
ENST00000545193.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr17_+_57642886 | 0.10 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr17_-_26903900 | 0.10 |
ENST00000395319.3
ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC
|
aldolase C, fructose-bisphosphate |
| chr4_-_105416039 | 0.10 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr9_+_131683174 | 0.10 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr19_+_41119794 | 0.10 |
ENST00000593463.1
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr2_-_165698521 | 0.10 |
ENST00000409184.3
ENST00000392717.2 ENST00000456693.1 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr4_-_71705060 | 0.10 |
ENST00000514161.1
|
GRSF1
|
G-rich RNA sequence binding factor 1 |
| chr2_-_179315453 | 0.10 |
ENST00000432031.2
|
PRKRA
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr2_-_165697717 | 0.10 |
ENST00000439313.1
|
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr17_-_26694979 | 0.09 |
ENST00000438614.1
|
VTN
|
vitronectin |
| chr6_+_167412835 | 0.09 |
ENST00000349556.4
|
FGFR1OP
|
FGFR1 oncogene partner |
| chr13_+_102104952 | 0.09 |
ENST00000376180.3
|
ITGBL1
|
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr6_-_32908792 | 0.09 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr2_+_74361599 | 0.09 |
ENST00000401851.1
|
MGC10955
|
MGC10955 |
| chr8_+_40010989 | 0.09 |
ENST00000315792.3
|
C8orf4
|
chromosome 8 open reading frame 4 |
| chr4_+_14113592 | 0.09 |
ENST00000502759.1
ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085
|
long intergenic non-protein coding RNA 1085 |
| chr12_+_50146747 | 0.09 |
ENST00000547798.1
|
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr11_-_113577052 | 0.09 |
ENST00000540540.1
ENST00000545579.1 ENST00000538955.1 ENST00000299882.5 |
TMPRSS5
|
transmembrane protease, serine 5 |
| chr7_-_129845313 | 0.09 |
ENST00000397622.2
|
TMEM209
|
transmembrane protein 209 |
| chr2_+_191045656 | 0.09 |
ENST00000443551.2
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr7_+_138145145 | 0.09 |
ENST00000415680.2
|
TRIM24
|
tripartite motif containing 24 |
| chr19_+_52264449 | 0.09 |
ENST00000599326.1
ENST00000598953.1 |
FPR2
|
formyl peptide receptor 2 |
| chr12_+_58003935 | 0.09 |
ENST00000333972.7
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25 |
| chr13_+_78315466 | 0.08 |
ENST00000314070.5
ENST00000462234.1 |
SLAIN1
|
SLAIN motif family, member 1 |
| chr20_+_33589048 | 0.08 |
ENST00000446156.1
ENST00000453028.1 ENST00000435272.1 ENST00000433934.2 ENST00000456649.1 |
MYH7B
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr17_+_46970127 | 0.08 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_+_22022437 | 0.08 |
ENST00000540040.1
|
MTRNR2L1
|
MT-RNR2-like 1 |
| chr8_+_82644669 | 0.08 |
ENST00000297265.4
|
CHMP4C
|
charged multivesicular body protein 4C |
| chr2_-_188312971 | 0.08 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr3_+_29323043 | 0.08 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr16_+_50582222 | 0.08 |
ENST00000268459.3
|
NKD1
|
naked cuticle homolog 1 (Drosophila) |
| chr8_+_35649365 | 0.08 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr22_+_38219389 | 0.08 |
ENST00000249041.2
|
GALR3
|
galanin receptor 3 |
| chr17_+_73997796 | 0.08 |
ENST00000586261.1
|
CDK3
|
cyclin-dependent kinase 3 |
| chr12_-_71551652 | 0.08 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr3_+_137906109 | 0.08 |
ENST00000481646.1
ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr8_+_10530155 | 0.08 |
ENST00000521818.1
|
C8orf74
|
chromosome 8 open reading frame 74 |
| chrX_+_99899180 | 0.08 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr5_+_52776449 | 0.08 |
ENST00000396947.3
|
FST
|
follistatin |
| chrX_-_48328631 | 0.08 |
ENST00000429543.1
ENST00000317669.5 |
SLC38A5
|
solute carrier family 38, member 5 |
| chr6_-_88299678 | 0.07 |
ENST00000369536.5
|
RARS2
|
arginyl-tRNA synthetase 2, mitochondrial |
| chr3_+_125985620 | 0.07 |
ENST00000511512.1
ENST00000512435.1 |
RP11-71E19.1
|
RP11-71E19.1 |
| chr7_-_129845188 | 0.07 |
ENST00000462753.1
ENST00000471077.1 ENST00000473456.1 ENST00000336804.8 |
TMEM209
|
transmembrane protein 209 |
| chr15_+_93447675 | 0.07 |
ENST00000536619.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr3_+_137906353 | 0.07 |
ENST00000461822.1
ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8
|
armadillo repeat containing 8 |
| chr20_+_23016057 | 0.07 |
ENST00000255008.3
|
SSTR4
|
somatostatin receptor 4 |
| chr2_+_130737223 | 0.07 |
ENST00000410061.2
|
RAB6C
|
RAB6C, member RAS oncogene family |
| chr1_+_155829341 | 0.07 |
ENST00000539162.1
|
SYT11
|
synaptotagmin XI |
| chr7_+_73442487 | 0.07 |
ENST00000380575.4
ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN
|
elastin |
| chr17_+_46970178 | 0.07 |
ENST00000393366.2
ENST00000506855.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr10_-_114206649 | 0.07 |
ENST00000369404.3
ENST00000369405.3 |
ZDHHC6
|
zinc finger, DHHC-type containing 6 |
| chr5_+_102201687 | 0.07 |
ENST00000304400.7
|
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_-_61244308 | 0.07 |
ENST00000407787.1
ENST00000398658.2 |
PUS10
|
pseudouridylate synthase 10 |
| chrX_+_109602039 | 0.07 |
ENST00000520821.1
|
RGAG1
|
retrotransposon gag domain containing 1 |
| chr7_+_130126165 | 0.07 |
ENST00000427521.1
ENST00000416162.2 ENST00000378576.4 |
MEST
|
mesoderm specific transcript |
| chr10_+_94352956 | 0.07 |
ENST00000260731.3
|
KIF11
|
kinesin family member 11 |
| chr4_+_177241094 | 0.07 |
ENST00000503362.1
|
SPCS3
|
signal peptidase complex subunit 3 homolog (S. cerevisiae) |
| chr12_+_7456880 | 0.07 |
ENST00000399422.4
|
ACSM4
|
acyl-CoA synthetase medium-chain family member 4 |
| chrX_-_48328551 | 0.06 |
ENST00000376876.3
|
SLC38A5
|
solute carrier family 38, member 5 |
| chr12_+_79258547 | 0.06 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr17_-_4463856 | 0.06 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr17_-_26697304 | 0.06 |
ENST00000536498.1
|
VTN
|
vitronectin |
| chr12_+_56473628 | 0.06 |
ENST00000549282.1
ENST00000549061.1 ENST00000267101.3 |
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr11_-_71810258 | 0.06 |
ENST00000544594.1
|
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr5_+_102201509 | 0.06 |
ENST00000348126.2
ENST00000379787.4 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chr2_-_183387430 | 0.06 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr11_-_10920714 | 0.06 |
ENST00000533941.1
|
CTD-2003C8.2
|
CTD-2003C8.2 |
| chr19_-_42947121 | 0.06 |
ENST00000601181.1
|
CXCL17
|
chemokine (C-X-C motif) ligand 17 |
| chrX_+_9880590 | 0.06 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr17_+_38119216 | 0.06 |
ENST00000301659.4
|
GSDMA
|
gasdermin A |
| chr18_+_29171689 | 0.06 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr7_-_37026108 | 0.06 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr2_-_158182105 | 0.06 |
ENST00000409925.1
|
ERMN
|
ermin, ERM-like protein |
| chr20_-_60573188 | 0.06 |
ENST00000474089.1
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr19_+_42254885 | 0.06 |
ENST00000595740.1
|
CEACAM6
|
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| chr8_-_26371608 | 0.06 |
ENST00000522362.2
|
PNMA2
|
paraneoplastic Ma antigen 2 |
| chr1_+_204494618 | 0.06 |
ENST00000367180.1
ENST00000391947.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr17_-_42988004 | 0.06 |
ENST00000586125.1
ENST00000591880.1 |
GFAP
|
glial fibrillary acidic protein |
| chr1_+_154975110 | 0.06 |
ENST00000535420.1
ENST00000368426.3 |
ZBTB7B
|
zinc finger and BTB domain containing 7B |
| chr1_+_202183200 | 0.05 |
ENST00000439764.2
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr1_+_192778161 | 0.05 |
ENST00000235382.5
|
RGS2
|
regulator of G-protein signaling 2, 24kDa |
| chr2_+_64681219 | 0.05 |
ENST00000238875.5
|
LGALSL
|
lectin, galactoside-binding-like |
| chr3_+_44626359 | 0.05 |
ENST00000412641.1
|
ZNF197
|
zinc finger protein 197 |
| chr7_+_130126012 | 0.05 |
ENST00000341441.5
|
MEST
|
mesoderm specific transcript |
| chr17_+_41150290 | 0.05 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr11_-_17229480 | 0.05 |
ENST00000532035.1
ENST00000540361.1 |
PIK3C2A
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr17_+_46970134 | 0.05 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr1_-_145039771 | 0.05 |
ENST00000493130.2
ENST00000532801.1 ENST00000478649.2 |
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr21_+_17791648 | 0.05 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_166944652 | 0.05 |
ENST00000528703.1
ENST00000525740.1 ENST00000529387.1 ENST00000469934.2 ENST00000529071.1 ENST00000526687.1 |
ILDR2
|
immunoglobulin-like domain containing receptor 2 |
| chr5_-_76935513 | 0.05 |
ENST00000306422.3
|
OTP
|
orthopedia homeobox |
| chr5_-_148442584 | 0.05 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PGR
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.2 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.2 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.2 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) |
| 0.0 | 0.1 | GO:2000588 | positive regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000588) |
| 0.0 | 0.2 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.2 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.2 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.5 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.2 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.1 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:0003409 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.4 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0060620 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.1 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.4 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:1904714 | chaperone-mediated autophagy(GO:0061684) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.0 | 0.0 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) B cell cytokine production(GO:0002368) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.0 | GO:0060699 | regulation of endoribonuclease activity(GO:0060699) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.0 | GO:1902256 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) glomerular endothelium development(GO:0072011) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.2 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.4 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.2 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.3 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.0 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |