Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
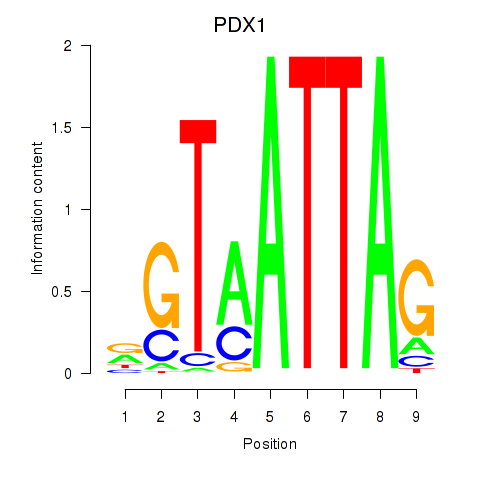
Results for PDX1
Z-value: 0.70
Transcription factors associated with PDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PDX1
|
ENSG00000139515.5 | pancreatic and duodenal homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PDX1 | hg19_v2_chr13_+_28494130_28494168 | -0.46 | 3.5e-01 | Click! |
Activity profile of PDX1 motif
Sorted Z-values of PDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_86650077 | 0.65 |
ENST00000552808.2
ENST00000547225.1 |
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr8_-_1922789 | 0.64 |
ENST00000521498.1
|
RP11-439C15.4
|
RP11-439C15.4 |
| chr22_-_28490123 | 0.56 |
ENST00000442232.1
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr4_+_53525573 | 0.52 |
ENST00000503051.1
|
USP46-AS1
|
USP46 antisense RNA 1 |
| chr1_+_62439037 | 0.50 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr11_+_115498761 | 0.49 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chr14_+_59100774 | 0.48 |
ENST00000556859.1
ENST00000421793.1 |
DACT1
|
dishevelled-binding antagonist of beta-catenin 1 |
| chr2_+_66918558 | 0.47 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr17_+_74846230 | 0.46 |
ENST00000592919.1
|
LINC00868
|
long intergenic non-protein coding RNA 868 |
| chr14_+_39944025 | 0.43 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr14_+_100485712 | 0.43 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr14_-_77889860 | 0.39 |
ENST00000555603.1
|
NOXRED1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr3_-_112127981 | 0.37 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr17_-_8301132 | 0.36 |
ENST00000399398.2
|
RNF222
|
ring finger protein 222 |
| chr1_-_159832438 | 0.35 |
ENST00000368100.1
|
VSIG8
|
V-set and immunoglobulin domain containing 8 |
| chr4_+_22999152 | 0.35 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr2_+_45168875 | 0.34 |
ENST00000260653.3
|
SIX3
|
SIX homeobox 3 |
| chr11_-_8285405 | 0.34 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr1_-_219615984 | 0.32 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr5_+_66300464 | 0.32 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr15_+_59910132 | 0.31 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr15_+_75080883 | 0.28 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chr12_+_107712173 | 0.28 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chrX_+_10031499 | 0.28 |
ENST00000454666.1
|
WWC3
|
WWC family member 3 |
| chr6_-_116833500 | 0.27 |
ENST00000356128.4
|
TRAPPC3L
|
trafficking protein particle complex 3-like |
| chr17_-_2117600 | 0.27 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr7_-_99716914 | 0.26 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr13_-_46716969 | 0.26 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr17_-_77924627 | 0.26 |
ENST00000572862.1
ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16
|
TBC1 domain family, member 16 |
| chr20_-_50418947 | 0.25 |
ENST00000371539.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr9_+_131037623 | 0.25 |
ENST00000495313.1
ENST00000372898.2 |
SWI5
|
SWI5 recombination repair homolog (yeast) |
| chr20_+_60174827 | 0.25 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr4_+_169013666 | 0.24 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr7_-_111424462 | 0.23 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr2_-_112237835 | 0.22 |
ENST00000442293.1
ENST00000439494.1 |
MIR4435-1HG
|
MIR4435-1 host gene (non-protein coding) |
| chr6_-_31107127 | 0.22 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr8_+_77593474 | 0.22 |
ENST00000455469.2
ENST00000050961.6 |
ZFHX4
|
zinc finger homeobox 4 |
| chr10_+_24497704 | 0.22 |
ENST00000376456.4
ENST00000458595.1 |
KIAA1217
|
KIAA1217 |
| chr2_-_8715616 | 0.21 |
ENST00000418358.1
|
AC011747.3
|
AC011747.3 |
| chr8_-_37594944 | 0.21 |
ENST00000330539.1
|
RP11-863K10.7
|
Uncharacterized protein |
| chr4_+_146402346 | 0.21 |
ENST00000514778.1
ENST00000507594.1 |
SMAD1
|
SMAD family member 1 |
| chr15_+_62853562 | 0.21 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr8_+_9953061 | 0.21 |
ENST00000522907.1
ENST00000528246.1 |
MSRA
|
methionine sulfoxide reductase A |
| chr14_-_92247032 | 0.21 |
ENST00000556661.1
ENST00000553676.1 ENST00000554560.1 |
CATSPERB
|
catsper channel auxiliary subunit beta |
| chr17_-_27467418 | 0.21 |
ENST00000528564.1
|
MYO18A
|
myosin XVIIIA |
| chr5_+_172068232 | 0.20 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chrX_+_10126488 | 0.19 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr7_+_114055052 | 0.18 |
ENST00000462331.1
ENST00000408937.3 ENST00000403559.4 ENST00000350908.4 ENST00000393498.2 ENST00000393495.3 ENST00000378237.3 ENST00000393489.3 |
FOXP2
|
forkhead box P2 |
| chr7_+_129015484 | 0.18 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr7_-_111424506 | 0.18 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr15_-_37393406 | 0.18 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr17_+_72427477 | 0.17 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr16_+_53133070 | 0.17 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr4_-_139163491 | 0.17 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr19_-_56110859 | 0.17 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr9_-_13165457 | 0.17 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr14_+_61654271 | 0.17 |
ENST00000555185.1
ENST00000557294.1 ENST00000556778.1 |
PRKCH
|
protein kinase C, eta |
| chr2_+_87769459 | 0.16 |
ENST00000414030.1
ENST00000437561.1 |
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chrX_-_21676442 | 0.16 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr8_+_77593448 | 0.16 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr8_+_9953214 | 0.16 |
ENST00000382490.5
|
MSRA
|
methionine sulfoxide reductase A |
| chr5_+_137722255 | 0.16 |
ENST00000542866.1
|
KDM3B
|
lysine (K)-specific demethylase 3B |
| chr14_-_95236551 | 0.16 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr20_-_50419055 | 0.16 |
ENST00000217086.4
|
SALL4
|
spalt-like transcription factor 4 |
| chr20_-_56286479 | 0.15 |
ENST00000265626.4
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr3_+_69985734 | 0.15 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr1_+_28586006 | 0.15 |
ENST00000253063.3
|
SESN2
|
sestrin 2 |
| chr17_-_18430160 | 0.15 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr15_+_59279851 | 0.15 |
ENST00000348370.4
ENST00000434298.1 ENST00000559160.1 |
RNF111
|
ring finger protein 111 |
| chr15_+_41913690 | 0.14 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr17_-_71223839 | 0.14 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr1_+_212965170 | 0.14 |
ENST00000532324.1
ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3
|
TatD DNase domain containing 3 |
| chr16_-_30457048 | 0.14 |
ENST00000500504.2
ENST00000542752.1 |
SEPHS2
|
selenophosphate synthetase 2 |
| chr8_+_22844995 | 0.14 |
ENST00000524077.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chrX_-_100129128 | 0.14 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr18_-_46784778 | 0.14 |
ENST00000582399.1
|
DYM
|
dymeclin |
| chr7_+_100860949 | 0.13 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr12_-_56321397 | 0.13 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr4_-_103749179 | 0.13 |
ENST00000502690.1
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_-_36301984 | 0.13 |
ENST00000502994.1
ENST00000515759.1 ENST00000296604.3 |
RANBP3L
|
RAN binding protein 3-like |
| chr11_-_129062093 | 0.13 |
ENST00000310343.9
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr6_+_13272904 | 0.12 |
ENST00000379335.3
ENST00000379329.1 |
PHACTR1
|
phosphatase and actin regulator 1 |
| chr1_+_198608146 | 0.12 |
ENST00000367376.2
ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC
|
protein tyrosine phosphatase, receptor type, C |
| chr13_-_99667960 | 0.12 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr10_+_24738355 | 0.12 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr11_+_130318869 | 0.12 |
ENST00000299164.2
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
| chr3_-_98235962 | 0.12 |
ENST00000513873.1
|
CLDND1
|
claudin domain containing 1 |
| chr4_-_41884620 | 0.12 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr1_+_19967014 | 0.12 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr5_+_98109322 | 0.12 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr5_-_140998481 | 0.11 |
ENST00000518047.1
|
DIAPH1
|
diaphanous-related formin 1 |
| chr17_-_39623681 | 0.11 |
ENST00000225899.3
|
KRT32
|
keratin 32 |
| chr12_+_64798826 | 0.11 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr8_+_38831683 | 0.10 |
ENST00000302495.4
|
HTRA4
|
HtrA serine peptidase 4 |
| chr9_-_117267717 | 0.10 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr9_-_123639600 | 0.10 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr6_-_33160231 | 0.10 |
ENST00000395194.1
ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2
|
collagen, type XI, alpha 2 |
| chr7_-_100860851 | 0.10 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chrX_+_7137475 | 0.10 |
ENST00000217961.4
|
STS
|
steroid sulfatase (microsomal), isozyme S |
| chr17_-_33446735 | 0.10 |
ENST00000460118.2
ENST00000335858.7 |
RAD51D
|
RAD51 paralog D |
| chr7_-_99717463 | 0.10 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr5_+_66300446 | 0.10 |
ENST00000261569.7
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_94147385 | 0.09 |
ENST00000260502.6
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr2_+_109204743 | 0.09 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_-_119759795 | 0.09 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr2_+_96331830 | 0.09 |
ENST00000425887.2
|
AC008268.1
|
AC008268.1 |
| chr4_+_141445333 | 0.09 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr4_+_144354644 | 0.09 |
ENST00000512843.1
|
GAB1
|
GRB2-associated binding protein 1 |
| chr3_+_69985792 | 0.09 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr5_-_140998616 | 0.09 |
ENST00000389054.3
ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1
|
diaphanous-related formin 1 |
| chr15_+_69854027 | 0.09 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr2_-_208031943 | 0.09 |
ENST00000421199.1
ENST00000457962.1 |
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_-_232651312 | 0.09 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr14_-_69261310 | 0.09 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr1_-_151431647 | 0.08 |
ENST00000368863.2
ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ
|
pogo transposable element with ZNF domain |
| chr10_-_21806759 | 0.08 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr15_-_72563585 | 0.08 |
ENST00000287196.9
ENST00000260376.7 |
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr1_-_1710229 | 0.08 |
ENST00000341991.3
|
NADK
|
NAD kinase |
| chr13_-_67802549 | 0.08 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr20_-_31124186 | 0.08 |
ENST00000375678.3
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr12_-_56321649 | 0.08 |
ENST00000454792.2
ENST00000408946.2 |
WIBG
|
within bgcn homolog (Drosophila) |
| chrX_-_19817869 | 0.08 |
ENST00000379698.4
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr7_+_129015671 | 0.08 |
ENST00000466993.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr8_+_22844913 | 0.08 |
ENST00000519685.1
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr4_+_77356248 | 0.08 |
ENST00000296043.6
|
SHROOM3
|
shroom family member 3 |
| chr15_-_34875771 | 0.08 |
ENST00000267731.7
|
GOLGA8B
|
golgin A8 family, member B |
| chr20_-_271304 | 0.08 |
ENST00000400269.3
ENST00000360321.2 |
C20orf96
|
chromosome 20 open reading frame 96 |
| chr1_-_151431909 | 0.08 |
ENST00000361398.3
ENST00000271715.2 |
POGZ
|
pogo transposable element with ZNF domain |
| chr20_+_43538692 | 0.08 |
ENST00000217074.4
ENST00000255136.3 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr17_-_9929581 | 0.08 |
ENST00000437099.2
ENST00000396115.2 |
GAS7
|
growth arrest-specific 7 |
| chr2_+_191208656 | 0.07 |
ENST00000458647.1
|
INPP1
|
inositol polyphosphate-1-phosphatase |
| chr19_-_12889226 | 0.07 |
ENST00000589400.1
ENST00000590839.1 ENST00000592079.1 |
HOOK2
|
hook microtubule-tethering protein 2 |
| chr11_-_31531121 | 0.07 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chrX_-_106146547 | 0.07 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr17_-_57229155 | 0.07 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr9_-_136933615 | 0.07 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr8_-_37457350 | 0.07 |
ENST00000519691.1
|
RP11-150O12.3
|
RP11-150O12.3 |
| chr15_-_55562479 | 0.07 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr1_-_1709845 | 0.07 |
ENST00000341426.5
ENST00000344463.4 |
NADK
|
NAD kinase |
| chr3_-_39321512 | 0.07 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr9_-_123639304 | 0.07 |
ENST00000436309.1
|
PHF19
|
PHD finger protein 19 |
| chr5_-_134735568 | 0.07 |
ENST00000510038.1
ENST00000304332.4 |
H2AFY
|
H2A histone family, member Y |
| chr5_+_174151536 | 0.07 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr7_+_116660246 | 0.06 |
ENST00000434836.1
ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr14_-_78083112 | 0.06 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr8_-_131399110 | 0.06 |
ENST00000521426.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr15_+_91416092 | 0.06 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr15_-_65407524 | 0.06 |
ENST00000559089.1
|
UBAP1L
|
ubiquitin associated protein 1-like |
| chr20_-_50418972 | 0.06 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr7_-_99716952 | 0.06 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr12_+_15699286 | 0.06 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr12_-_109221160 | 0.06 |
ENST00000326470.5
|
SSH1
|
slingshot protein phosphatase 1 |
| chr18_+_3447572 | 0.06 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr1_+_36621174 | 0.06 |
ENST00000429533.2
|
MAP7D1
|
MAP7 domain containing 1 |
| chr12_+_47610315 | 0.06 |
ENST00000548348.1
ENST00000549500.1 |
PCED1B
|
PC-esterase domain containing 1B |
| chr1_+_174844645 | 0.06 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chrX_-_13835147 | 0.06 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr4_+_71587669 | 0.06 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr3_-_121740969 | 0.05 |
ENST00000393631.1
ENST00000273691.3 ENST00000344209.5 |
ILDR1
|
immunoglobulin-like domain containing receptor 1 |
| chr5_-_142780280 | 0.05 |
ENST00000424646.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr15_-_100882191 | 0.05 |
ENST00000268070.4
|
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr4_-_109541610 | 0.05 |
ENST00000510212.1
|
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr5_-_115890554 | 0.05 |
ENST00000509665.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr11_-_85430163 | 0.05 |
ENST00000529581.1
ENST00000533577.1 |
SYTL2
|
synaptotagmin-like 2 |
| chrX_-_46187069 | 0.05 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr12_-_57328187 | 0.05 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr2_+_48541776 | 0.05 |
ENST00000413569.1
ENST00000340553.3 |
FOXN2
|
forkhead box N2 |
| chr4_+_151503077 | 0.05 |
ENST00000317605.4
|
MAB21L2
|
mab-21-like 2 (C. elegans) |
| chr15_+_28624878 | 0.05 |
ENST00000450328.2
|
GOLGA8F
|
golgin A8 family, member F |
| chr20_+_43538756 | 0.05 |
ENST00000537323.1
ENST00000217073.2 |
PABPC1L
|
poly(A) binding protein, cytoplasmic 1-like |
| chr1_-_151431674 | 0.05 |
ENST00000531094.1
|
POGZ
|
pogo transposable element with ZNF domain |
| chr9_-_123639445 | 0.04 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr8_+_22424551 | 0.04 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr12_-_86650045 | 0.04 |
ENST00000604798.1
|
MGAT4C
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr18_+_29027696 | 0.04 |
ENST00000257189.4
|
DSG3
|
desmoglein 3 |
| chr10_-_21186144 | 0.04 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr12_-_56106060 | 0.04 |
ENST00000452168.2
|
ITGA7
|
integrin, alpha 7 |
| chr15_+_67841330 | 0.04 |
ENST00000354498.5
|
MAP2K5
|
mitogen-activated protein kinase kinase 5 |
| chr1_-_2458026 | 0.04 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr1_-_234667504 | 0.04 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr12_-_51611477 | 0.04 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr15_+_93443419 | 0.04 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr1_+_151735431 | 0.04 |
ENST00000321531.5
ENST00000315067.8 |
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr3_+_35722487 | 0.04 |
ENST00000441454.1
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa |
| chr7_-_73038867 | 0.04 |
ENST00000313375.3
ENST00000354613.1 ENST00000395189.1 ENST00000453275.1 |
MLXIPL
|
MLX interacting protein-like |
| chr1_+_84609944 | 0.04 |
ENST00000370685.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_-_152147579 | 0.04 |
ENST00000304527.4
ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19
|
SH3 domain containing 19 |
| chr12_-_32908809 | 0.04 |
ENST00000324868.8
|
YARS2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr4_-_70518941 | 0.03 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr4_-_66536057 | 0.03 |
ENST00000273854.3
|
EPHA5
|
EPH receptor A5 |
| chr5_+_156712372 | 0.03 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr11_-_85430088 | 0.03 |
ENST00000533057.1
ENST00000533892.1 |
SYTL2
|
synaptotagmin-like 2 |
| chr9_-_136933134 | 0.03 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr20_+_43990576 | 0.03 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr7_-_99716940 | 0.03 |
ENST00000440225.1
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr15_-_55563072 | 0.03 |
ENST00000567380.1
ENST00000565972.1 ENST00000569493.1 |
RAB27A
|
RAB27A, member RAS oncogene family |
| chr18_+_32558208 | 0.03 |
ENST00000436190.2
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr6_+_43968306 | 0.03 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr9_+_131062367 | 0.03 |
ENST00000601297.1
|
AL359091.2
|
CDNA: FLJ21673 fis, clone COL09042; HCG2036511; Uncharacterized protein |
| chr3_-_141747950 | 0.03 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
Network of associatons between targets according to the STRING database.
First level regulatory network of PDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.1 | 0.3 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.1 | 0.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.5 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.4 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.2 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.2 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.3 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.4 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.1 | GO:1904815 | negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0032904 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0035701 | immunoglobulin biosynthetic process(GO:0002378) hematopoietic stem cell migration(GO:0035701) bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.2 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.3 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.5 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.0 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |