Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
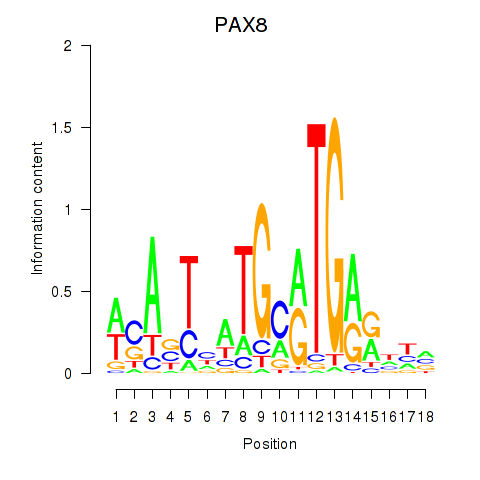
Results for PAX8
Z-value: 1.21
Transcription factors associated with PAX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX8
|
ENSG00000125618.12 | paired box 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX8 | hg19_v2_chr2_-_114036488_114036527 | 0.59 | 2.2e-01 | Click! |
Activity profile of PAX8 motif
Sorted Z-values of PAX8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_24126350 | 1.21 |
ENST00000378491.4
ENST00000378478.1 ENST00000378477.2 |
NRSN1
|
neurensin 1 |
| chr6_+_131571535 | 1.16 |
ENST00000474850.2
|
AKAP7
|
A kinase (PRKA) anchor protein 7 |
| chr14_-_57197224 | 1.02 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr15_-_29745003 | 0.90 |
ENST00000560082.1
|
FAM189A1
|
family with sequence similarity 189, member A1 |
| chr11_-_9482010 | 0.88 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr19_+_35630926 | 0.87 |
ENST00000588081.1
ENST00000589121.1 |
FXYD1
|
FXYD domain containing ion transport regulator 1 |
| chrX_-_153141783 | 0.85 |
ENST00000458029.1
|
L1CAM
|
L1 cell adhesion molecule |
| chr22_-_20138302 | 0.72 |
ENST00000540078.1
ENST00000439765.2 |
AC006547.14
|
uncharacterized protein LOC388849 |
| chr4_+_123653807 | 0.72 |
ENST00000314218.3
ENST00000542236.1 |
BBS12
|
Bardet-Biedl syndrome 12 |
| chr9_-_116837249 | 0.66 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr2_-_87248975 | 0.65 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr4_+_123653882 | 0.63 |
ENST00000433287.1
|
BBS12
|
Bardet-Biedl syndrome 12 |
| chr1_-_119683251 | 0.62 |
ENST00000369426.5
ENST00000235521.4 |
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr12_+_72058130 | 0.62 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr19_-_39303576 | 0.58 |
ENST00000594209.1
|
LGALS4
|
lectin, galactoside-binding, soluble, 4 |
| chr10_+_85933494 | 0.58 |
ENST00000372126.3
|
C10orf99
|
chromosome 10 open reading frame 99 |
| chr1_+_203734296 | 0.58 |
ENST00000442561.2
ENST00000367217.5 |
LAX1
|
lymphocyte transmembrane adaptor 1 |
| chr6_+_8652370 | 0.58 |
ENST00000503668.1
|
HULC
|
hepatocellular carcinoma up-regulated long non-coding RNA |
| chr8_+_86851932 | 0.57 |
ENST00000517368.1
|
CTA-392E5.1
|
CTA-392E5.1 |
| chr16_+_75690093 | 0.57 |
ENST00000564671.2
|
TERF2IP
|
telomeric repeat binding factor 2, interacting protein |
| chr1_-_119682812 | 0.56 |
ENST00000537870.1
|
WARS2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| chr12_+_57828521 | 0.55 |
ENST00000309668.2
|
INHBC
|
inhibin, beta C |
| chr19_+_9361606 | 0.47 |
ENST00000456448.1
|
OR7E24
|
olfactory receptor, family 7, subfamily E, member 24 |
| chr19_+_52901094 | 0.46 |
ENST00000391788.2
ENST00000436397.1 ENST00000391787.2 ENST00000360465.3 ENST00000494167.2 ENST00000493272.1 |
ZNF528
|
zinc finger protein 528 |
| chr4_-_70653673 | 0.46 |
ENST00000512870.1
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr1_+_16083098 | 0.45 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr19_+_42041860 | 0.45 |
ENST00000483481.2
ENST00000494375.2 |
AC006129.4
|
AC006129.4 |
| chr3_-_137851220 | 0.45 |
ENST00000236709.3
|
A4GNT
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr21_-_44582378 | 0.44 |
ENST00000433840.1
|
AP001631.10
|
AP001631.10 |
| chr17_-_1508379 | 0.43 |
ENST00000412517.3
|
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr13_+_27844464 | 0.41 |
ENST00000241463.4
|
RASL11A
|
RAS-like, family 11, member A |
| chr4_+_154073469 | 0.36 |
ENST00000441616.1
|
TRIM2
|
tripartite motif containing 2 |
| chr8_-_66474884 | 0.36 |
ENST00000520902.1
|
CTD-3025N20.2
|
CTD-3025N20.2 |
| chr6_+_63921399 | 0.35 |
ENST00000356170.3
|
FKBP1C
|
FK506 binding protein 1C |
| chr2_+_38177575 | 0.33 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr11_+_20044375 | 0.32 |
ENST00000525322.1
ENST00000530408.1 |
NAV2
|
neuron navigator 2 |
| chr1_+_100817262 | 0.32 |
ENST00000455467.1
|
CDC14A
|
cell division cycle 14A |
| chr3_-_8811288 | 0.31 |
ENST00000316793.3
ENST00000431493.1 |
OXTR
|
oxytocin receptor |
| chr7_+_129984630 | 0.31 |
ENST00000355388.3
ENST00000497503.1 ENST00000463587.1 ENST00000461828.1 ENST00000494311.1 ENST00000466363.2 ENST00000485477.1 ENST00000431780.2 ENST00000474905.1 |
CPA5
|
carboxypeptidase A5 |
| chr6_+_160542870 | 0.31 |
ENST00000324965.4
ENST00000457470.2 |
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr1_-_21059029 | 0.30 |
ENST00000444387.2
ENST00000375031.1 ENST00000518294.1 |
SH2D5
|
SH2 domain containing 5 |
| chr17_-_39216344 | 0.30 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr19_-_35992780 | 0.30 |
ENST00000593342.1
ENST00000601650.1 ENST00000408915.2 |
DMKN
|
dermokine |
| chr6_+_161123270 | 0.29 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
| chr6_+_135502501 | 0.29 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr5_+_148521136 | 0.29 |
ENST00000506113.1
|
ABLIM3
|
actin binding LIM protein family, member 3 |
| chr8_-_57233103 | 0.28 |
ENST00000303749.3
ENST00000522671.1 |
SDR16C5
|
short chain dehydrogenase/reductase family 16C, member 5 |
| chr1_-_52520828 | 0.28 |
ENST00000610127.1
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr6_+_135502408 | 0.27 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr17_-_72588422 | 0.27 |
ENST00000375352.1
|
CD300LD
|
CD300 molecule-like family member d |
| chr4_-_23735183 | 0.27 |
ENST00000507666.1
|
RP11-380P13.2
|
RP11-380P13.2 |
| chr17_-_60883993 | 0.26 |
ENST00000583803.1
ENST00000456609.2 |
MARCH10
|
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr19_+_41856816 | 0.25 |
ENST00000539627.1
|
TMEM91
|
transmembrane protein 91 |
| chr11_+_20044096 | 0.24 |
ENST00000533917.1
|
NAV2
|
neuron navigator 2 |
| chr21_+_45287112 | 0.24 |
ENST00000448287.1
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr11_+_107461948 | 0.23 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr19_+_50169216 | 0.23 |
ENST00000594157.1
ENST00000600947.1 ENST00000598306.1 |
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr12_-_92821922 | 0.23 |
ENST00000538965.1
ENST00000378487.2 |
CLLU1OS
|
chronic lymphocytic leukemia up-regulated 1 opposite strand |
| chr11_+_117857063 | 0.22 |
ENST00000227752.3
ENST00000541785.1 ENST00000545409.1 |
IL10RA
|
interleukin 10 receptor, alpha |
| chr12_-_129308487 | 0.22 |
ENST00000266771.5
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr11_-_118272610 | 0.22 |
ENST00000534438.1
|
RP11-770J1.5
|
Uncharacterized protein |
| chr15_+_34638066 | 0.22 |
ENST00000333756.4
|
NUTM1
|
NUT midline carcinoma, family member 1 |
| chr4_+_72204755 | 0.22 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr1_+_161494036 | 0.21 |
ENST00000309758.4
|
HSPA6
|
heat shock 70kDa protein 6 (HSP70B') |
| chr7_+_140396756 | 0.21 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr12_+_26348582 | 0.21 |
ENST00000535504.1
|
SSPN
|
sarcospan |
| chr9_+_131549610 | 0.21 |
ENST00000223865.8
|
TBC1D13
|
TBC1 domain family, member 13 |
| chr4_-_69536346 | 0.20 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr2_+_219524473 | 0.20 |
ENST00000439945.1
ENST00000431802.1 |
BCS1L
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr6_+_151773583 | 0.20 |
ENST00000545879.1
|
C6orf211
|
chromosome 6 open reading frame 211 |
| chr11_+_20044600 | 0.20 |
ENST00000311043.8
|
NAV2
|
neuron navigator 2 |
| chr10_+_12391685 | 0.20 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr13_+_51483814 | 0.19 |
ENST00000336617.3
ENST00000422660.1 |
RNASEH2B
|
ribonuclease H2, subunit B |
| chr12_-_123187890 | 0.19 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr3_+_186435065 | 0.19 |
ENST00000287611.2
ENST00000265023.4 |
KNG1
|
kininogen 1 |
| chr9_-_98269481 | 0.18 |
ENST00000418258.1
ENST00000553011.1 ENST00000551845.1 |
PTCH1
|
patched 1 |
| chrX_-_151903184 | 0.18 |
ENST00000357916.4
ENST00000393869.3 |
MAGEA12
|
melanoma antigen family A, 12 |
| chr9_-_128246769 | 0.18 |
ENST00000444226.1
|
MAPKAP1
|
mitogen-activated protein kinase associated protein 1 |
| chr1_+_155006300 | 0.18 |
ENST00000295542.1
ENST00000392480.1 ENST00000423025.2 ENST00000368419.2 |
DCST1
|
DC-STAMP domain containing 1 |
| chr9_-_92020841 | 0.18 |
ENST00000433650.1
|
SEMA4D
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr15_+_23810853 | 0.17 |
ENST00000568252.1
|
MKRN3
|
makorin ring finger protein 3 |
| chr19_-_37701386 | 0.17 |
ENST00000527838.1
ENST00000591492.1 ENST00000532828.2 |
ZNF585B
|
zinc finger protein 585B |
| chrX_-_153141434 | 0.16 |
ENST00000407935.2
ENST00000439496.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr6_+_41196077 | 0.16 |
ENST00000448827.2
|
TREML4
|
triggering receptor expressed on myeloid cells-like 4 |
| chrX_-_153141302 | 0.16 |
ENST00000361699.4
ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM
|
L1 cell adhesion molecule |
| chr19_+_35842445 | 0.16 |
ENST00000246553.2
|
FFAR1
|
free fatty acid receptor 1 |
| chr18_-_58040000 | 0.16 |
ENST00000299766.3
|
MC4R
|
melanocortin 4 receptor |
| chr7_+_140396946 | 0.16 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr14_+_58797974 | 0.16 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr9_+_71944241 | 0.16 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr16_-_31519691 | 0.16 |
ENST00000567994.1
ENST00000430477.2 ENST00000570164.1 ENST00000327237.2 |
C16orf58
|
chromosome 16 open reading frame 58 |
| chr7_+_140396465 | 0.16 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr11_+_59705928 | 0.15 |
ENST00000398992.1
|
OOSP1
|
oocyte secreted protein 1 |
| chr2_+_85661918 | 0.15 |
ENST00000340326.2
|
SH2D6
|
SH2 domain containing 6 |
| chr16_-_15736345 | 0.15 |
ENST00000549219.1
|
KIAA0430
|
KIAA0430 |
| chr7_+_90012986 | 0.15 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr17_+_6918093 | 0.15 |
ENST00000439424.2
|
C17orf49
|
chromosome 17 open reading frame 49 |
| chr6_+_135502466 | 0.14 |
ENST00000367814.4
|
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr7_-_128001658 | 0.14 |
ENST00000489835.2
ENST00000464607.1 ENST00000489517.1 ENST00000446477.2 ENST00000535159.1 ENST00000435512.1 ENST00000495931.1 |
PRRT4
|
proline-rich transmembrane protein 4 |
| chr12_-_11287243 | 0.14 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr12_+_133757995 | 0.14 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr15_+_75303993 | 0.14 |
ENST00000564779.1
|
SCAMP5
|
secretory carrier membrane protein 5 |
| chr6_+_43266063 | 0.14 |
ENST00000372574.3
|
SLC22A7
|
solute carrier family 22 (organic anion transporter), member 7 |
| chr11_-_104905840 | 0.14 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr3_+_44754126 | 0.13 |
ENST00000449836.1
ENST00000436624.2 ENST00000296091.4 ENST00000411443.1 |
ZNF502
|
zinc finger protein 502 |
| chr19_+_782755 | 0.13 |
ENST00000606242.1
ENST00000586061.1 |
AC006273.5
|
AC006273.5 |
| chr5_-_156390230 | 0.13 |
ENST00000407087.3
ENST00000274532.2 |
TIMD4
|
T-cell immunoglobulin and mucin domain containing 4 |
| chr15_+_89631647 | 0.13 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr4_-_128887069 | 0.13 |
ENST00000541133.1
ENST00000296468.3 ENST00000513559.1 |
MFSD8
|
major facilitator superfamily domain containing 8 |
| chr12_+_113229962 | 0.13 |
ENST00000553114.1
ENST00000548866.1 |
RPH3A
|
rabphilin 3A homolog (mouse) |
| chr18_+_6256746 | 0.13 |
ENST00000578427.1
|
RP11-760N9.1
|
RP11-760N9.1 |
| chr1_+_16083123 | 0.13 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr13_-_24471194 | 0.13 |
ENST00000382137.3
ENST00000382057.3 |
C1QTNF9B
|
C1q and tumor necrosis factor related protein 9B |
| chr6_+_160542821 | 0.13 |
ENST00000366963.4
|
SLC22A1
|
solute carrier family 22 (organic cation transporter), member 1 |
| chr12_-_6960407 | 0.12 |
ENST00000540683.1
ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3
|
cell division cycle associated 3 |
| chr5_-_140027357 | 0.12 |
ENST00000252102.4
|
NDUFA2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr11_+_113185251 | 0.12 |
ENST00000529221.1
|
TTC12
|
tetratricopeptide repeat domain 12 |
| chr4_+_71600063 | 0.12 |
ENST00000513597.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr6_-_27858570 | 0.12 |
ENST00000359303.2
|
HIST1H3J
|
histone cluster 1, H3j |
| chr6_-_44233361 | 0.12 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr9_-_117853297 | 0.12 |
ENST00000542877.1
ENST00000537320.1 ENST00000341037.4 |
TNC
|
tenascin C |
| chr15_-_49255632 | 0.12 |
ENST00000332408.4
|
SHC4
|
SHC (Src homology 2 domain containing) family, member 4 |
| chr11_-_58343319 | 0.12 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr1_+_28099700 | 0.12 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr19_+_29097595 | 0.11 |
ENST00000587961.1
|
AC079466.1
|
AC079466.1 |
| chr2_+_120301997 | 0.11 |
ENST00000602047.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr11_+_63742050 | 0.11 |
ENST00000314133.3
ENST00000535431.1 |
COX8A
AP000721.4
|
cytochrome c oxidase subunit VIIIA (ubiquitous) Uncharacterized protein |
| chr17_-_73266616 | 0.11 |
ENST00000579194.1
ENST00000581777.1 |
MIF4GD
|
MIF4G domain containing |
| chr9_-_134955246 | 0.11 |
ENST00000357028.2
ENST00000474263.1 ENST00000292035.5 |
MED27
|
mediator complex subunit 27 |
| chr3_+_11178779 | 0.11 |
ENST00000438284.2
|
HRH1
|
histamine receptor H1 |
| chr3_-_15540055 | 0.11 |
ENST00000605797.1
ENST00000435459.2 |
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr11_+_82904858 | 0.11 |
ENST00000260047.6
|
ANKRD42
|
ankyrin repeat domain 42 |
| chr4_-_70361579 | 0.11 |
ENST00000512583.1
|
UGT2B4
|
UDP glucuronosyltransferase 2 family, polypeptide B4 |
| chr9_+_124048864 | 0.11 |
ENST00000545652.1
|
GSN
|
gelsolin |
| chr7_+_149535455 | 0.11 |
ENST00000223210.4
ENST00000460379.1 |
ZNF862
|
zinc finger protein 862 |
| chr17_+_7461781 | 0.11 |
ENST00000349228.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr4_+_118955500 | 0.10 |
ENST00000296499.5
|
NDST3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr19_-_55791563 | 0.10 |
ENST00000588971.1
ENST00000255631.5 ENST00000587551.1 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr10_+_24738355 | 0.10 |
ENST00000307544.6
|
KIAA1217
|
KIAA1217 |
| chr19_-_55791540 | 0.10 |
ENST00000433386.2
|
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr7_+_150413645 | 0.10 |
ENST00000307194.5
|
GIMAP1
|
GTPase, IMAP family member 1 |
| chr19_+_54704718 | 0.10 |
ENST00000391752.1
ENST00000402367.1 ENST00000391751.3 |
RPS9
|
ribosomal protein S9 |
| chr12_+_122516626 | 0.10 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chrX_+_151867214 | 0.10 |
ENST00000329342.5
ENST00000412733.1 ENST00000457643.1 |
MAGEA6
|
melanoma antigen family A, 6 |
| chr17_+_7462103 | 0.10 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr19_+_54704610 | 0.10 |
ENST00000302907.4
|
RPS9
|
ribosomal protein S9 |
| chr1_-_85156417 | 0.10 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr1_+_40915725 | 0.10 |
ENST00000484445.1
ENST00000411995.2 ENST00000361584.3 |
ZFP69B
|
ZFP69 zinc finger protein B |
| chr17_-_73267281 | 0.09 |
ENST00000578305.1
ENST00000325102.8 |
MIF4GD
|
MIF4G domain containing |
| chr2_-_106013400 | 0.09 |
ENST00000409807.1
|
FHL2
|
four and a half LIM domains 2 |
| chr4_+_71600144 | 0.09 |
ENST00000502653.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr6_+_142623758 | 0.09 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr19_-_7812446 | 0.09 |
ENST00000394173.4
ENST00000301357.8 |
CD209
|
CD209 molecule |
| chr17_-_42100474 | 0.09 |
ENST00000585950.1
ENST00000592127.1 ENST00000589334.1 |
TMEM101
|
transmembrane protein 101 |
| chr17_-_73781567 | 0.09 |
ENST00000586607.1
|
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr16_+_66442411 | 0.09 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr17_+_7461849 | 0.09 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr4_-_70361615 | 0.09 |
ENST00000305107.6
|
UGT2B4
|
UDP glucuronosyltransferase 2 family, polypeptide B4 |
| chr3_-_78719376 | 0.09 |
ENST00000495961.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr19_-_36019123 | 0.09 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr7_+_6713376 | 0.09 |
ENST00000399484.3
ENST00000544825.1 ENST00000401847.1 |
AC073343.1
|
Uncharacterized protein |
| chr10_+_135340859 | 0.09 |
ENST00000252945.3
ENST00000421586.1 ENST00000418356.1 |
CYP2E1
|
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr10_+_35484793 | 0.09 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr21_-_43786634 | 0.09 |
ENST00000291527.2
|
TFF1
|
trefoil factor 1 |
| chr10_+_24544249 | 0.08 |
ENST00000430453.2
|
KIAA1217
|
KIAA1217 |
| chr1_+_44889697 | 0.08 |
ENST00000443020.2
|
RNF220
|
ring finger protein 220 |
| chr15_+_89631381 | 0.08 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chrY_-_20935572 | 0.08 |
ENST00000382852.1
ENST00000344884.4 ENST00000304790.3 |
HSFY2
|
heat shock transcription factor, Y linked 2 |
| chr21_-_43816052 | 0.08 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr19_-_7812397 | 0.08 |
ENST00000593660.1
ENST00000354397.6 ENST00000593821.1 ENST00000602261.1 ENST00000315591.8 ENST00000394161.5 ENST00000204801.8 ENST00000601256.1 ENST00000601951.1 ENST00000315599.7 |
CD209
|
CD209 molecule |
| chr3_+_164924769 | 0.08 |
ENST00000494915.1
|
RP11-85M11.2
|
RP11-85M11.2 |
| chr2_-_106013207 | 0.08 |
ENST00000447958.1
|
FHL2
|
four and a half LIM domains 2 |
| chr4_+_2800291 | 0.08 |
ENST00000504294.1
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr3_-_150264272 | 0.08 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr19_+_35939154 | 0.08 |
ENST00000599180.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr17_-_73267214 | 0.08 |
ENST00000580717.1
ENST00000577542.1 ENST00000579612.1 ENST00000245551.5 |
MIF4GD
|
MIF4G domain containing |
| chr7_-_112430647 | 0.08 |
ENST00000312814.6
|
TMEM168
|
transmembrane protein 168 |
| chr7_-_97501706 | 0.08 |
ENST00000455086.1
ENST00000453600.1 |
ASNS
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr12_-_6961050 | 0.08 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr13_-_103426112 | 0.08 |
ENST00000376032.4
ENST00000376029.3 |
TEX30
|
testis expressed 30 |
| chr6_+_36853607 | 0.08 |
ENST00000480824.2
ENST00000355190.3 ENST00000373685.1 |
C6orf89
|
chromosome 6 open reading frame 89 |
| chr8_+_39792474 | 0.08 |
ENST00000502986.2
|
IDO2
|
indoleamine 2,3-dioxygenase 2 |
| chr19_+_9938562 | 0.07 |
ENST00000586895.1
ENST00000358666.3 ENST00000590068.1 ENST00000593087.1 |
UBL5
|
ubiquitin-like 5 |
| chr19_+_50169081 | 0.07 |
ENST00000246784.3
|
BCL2L12
|
BCL2-like 12 (proline rich) |
| chr1_-_173176452 | 0.07 |
ENST00000281834.3
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr7_-_144533074 | 0.07 |
ENST00000360057.3
ENST00000378099.3 |
TPK1
|
thiamin pyrophosphokinase 1 |
| chrY_+_20708557 | 0.07 |
ENST00000307393.2
ENST00000309834.4 ENST00000382856.2 |
HSFY1
|
heat shock transcription factor, Y-linked 1 |
| chr2_+_201242941 | 0.07 |
ENST00000449647.1
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr1_+_89990378 | 0.07 |
ENST00000449440.1
|
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr5_+_140235469 | 0.07 |
ENST00000506939.2
ENST00000307360.5 |
PCDHA10
|
protocadherin alpha 10 |
| chr20_-_1373682 | 0.07 |
ENST00000381724.3
|
FKBP1A
|
FK506 binding protein 1A, 12kDa |
| chr19_-_55791431 | 0.07 |
ENST00000593263.1
ENST00000376343.3 |
HSPBP1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr17_-_67057203 | 0.07 |
ENST00000340001.4
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr10_-_81203972 | 0.06 |
ENST00000372333.3
|
ZCCHC24
|
zinc finger, CCHC domain containing 24 |
| chr11_-_62609281 | 0.06 |
ENST00000525239.1
ENST00000538098.2 |
WDR74
|
WD repeat domain 74 |
| chr19_+_16296191 | 0.06 |
ENST00000589852.1
ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A
|
family with sequence similarity 32, member A |
| chr10_+_96443378 | 0.06 |
ENST00000285979.6
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr16_+_66413128 | 0.06 |
ENST00000563425.2
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr14_+_76452090 | 0.06 |
ENST00000314067.6
ENST00000238628.6 ENST00000556742.1 |
IFT43
|
intraflagellar transport 43 homolog (Chlamydomonas) |
| chr5_-_176943917 | 0.06 |
ENST00000330503.7
|
DDX41
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.2 | 1.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.9 | GO:0010734 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.3 | GO:0034059 | response to anoxia(GO:0034059) positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.6 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.8 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) vagus nerve development(GO:0021564) |
| 0.1 | 0.4 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.1 | 0.2 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.1 | 0.2 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.2 | GO:0046968 | peptide antigen transport(GO:0046968) |
| 0.0 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 1.3 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.2 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.2 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.2 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:2000523 | negative regulation of regulatory T cell differentiation(GO:0045590) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.2 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0001507 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.3 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.2 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.6 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.6 | GO:0048305 | immunoglobulin secretion(GO:0048305) |
| 0.0 | 0.4 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 1.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.0 | 0.0 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) positive regulation of gastric acid secretion(GO:0060454) response to capsazepine(GO:1901594) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.9 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.4 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0019862 | IgA binding(GO:0019862) |
| 0.2 | 1.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.4 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.3 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.1 | 0.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.2 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.6 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 1.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.3 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |