Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
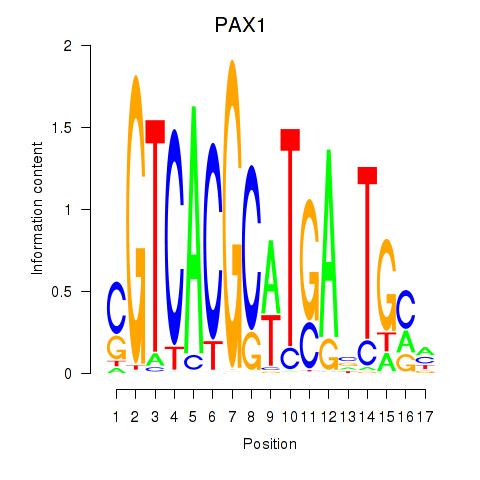
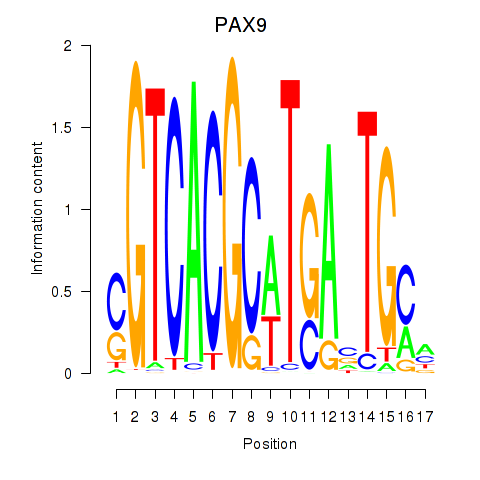
Results for PAX1_PAX9
Z-value: 0.40
Transcription factors associated with PAX1_PAX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX1
|
ENSG00000125813.9 | paired box 1 |
|
PAX9
|
ENSG00000198807.8 | paired box 9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX9 | hg19_v2_chr14_+_37131058_37131139 | -0.73 | 9.6e-02 | Click! |
| PAX1 | hg19_v2_chr20_+_21686290_21686311 | -0.15 | 7.8e-01 | Click! |
Activity profile of PAX1_PAX9 motif
Sorted Z-values of PAX1_PAX9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_121824374 | 0.42 |
ENST00000517992.1
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr19_-_409134 | 0.33 |
ENST00000332235.6
|
C2CD4C
|
C2 calcium-dependent domain containing 4C |
| chr18_-_47814032 | 0.22 |
ENST00000589548.1
ENST00000591474.1 |
CXXC1
|
CXXC finger protein 1 |
| chr19_-_18995029 | 0.18 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr9_+_33750515 | 0.13 |
ENST00000361005.5
|
PRSS3
|
protease, serine, 3 |
| chr17_-_3461092 | 0.13 |
ENST00000301365.4
ENST00000572519.1 |
TRPV3
|
transient receptor potential cation channel, subfamily V, member 3 |
| chr8_-_23282820 | 0.12 |
ENST00000520871.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr6_-_170893268 | 0.11 |
ENST00000538195.1
|
PDCD2
|
programmed cell death 2 |
| chr9_+_33750667 | 0.10 |
ENST00000457896.1
ENST00000342836.4 ENST00000429677.3 |
PRSS3
|
protease, serine, 3 |
| chr19_+_14184370 | 0.10 |
ENST00000590772.1
|
hsa-mir-1199
|
hsa-mir-1199 |
| chr14_-_69261310 | 0.09 |
ENST00000336440.3
|
ZFP36L1
|
ZFP36 ring finger protein-like 1 |
| chr8_-_23282797 | 0.09 |
ENST00000524144.1
|
LOXL2
|
lysyl oxidase-like 2 |
| chr20_-_17641097 | 0.09 |
ENST00000246043.4
|
RRBP1
|
ribosome binding protein 1 |
| chr1_-_204165610 | 0.09 |
ENST00000367194.4
|
KISS1
|
KiSS-1 metastasis-suppressor |
| chr18_-_47813940 | 0.09 |
ENST00000586837.1
ENST00000412036.2 ENST00000589940.1 |
CXXC1
|
CXXC finger protein 1 |
| chr1_+_26759295 | 0.07 |
ENST00000430232.1
|
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr11_-_67397371 | 0.07 |
ENST00000376693.2
ENST00000301490.4 |
NUDT8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr22_-_41940404 | 0.06 |
ENST00000355209.4
ENST00000337566.5 ENST00000396504.2 ENST00000407461.1 |
POLR3H
|
polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) |
| chr6_+_34857019 | 0.06 |
ENST00000360359.3
ENST00000535627.1 |
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr8_-_23261589 | 0.06 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chrX_-_153707545 | 0.05 |
ENST00000357360.4
|
LAGE3
|
L antigen family, member 3 |
| chr9_+_6716478 | 0.05 |
ENST00000452643.1
|
RP11-390F4.3
|
RP11-390F4.3 |
| chr19_-_40791211 | 0.05 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr2_-_162931052 | 0.05 |
ENST00000360534.3
|
DPP4
|
dipeptidyl-peptidase 4 |
| chr5_+_133842243 | 0.05 |
ENST00000515627.2
|
AC005355.2
|
AC005355.2 |
| chr17_-_7137582 | 0.04 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr2_+_234160340 | 0.04 |
ENST00000417017.1
ENST00000392020.4 ENST00000392018.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr9_+_33795533 | 0.04 |
ENST00000379405.3
|
PRSS3
|
protease, serine, 3 |
| chr14_+_102196739 | 0.04 |
ENST00000556973.1
|
RP11-796G6.2
|
Uncharacterized protein |
| chr22_+_39745930 | 0.04 |
ENST00000318801.4
ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1
|
synaptogyrin 1 |
| chr17_+_39968926 | 0.03 |
ENST00000585664.1
ENST00000585922.1 ENST00000429461.1 |
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr3_-_57113281 | 0.03 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr17_+_15848231 | 0.03 |
ENST00000304222.2
|
ADORA2B
|
adenosine A2b receptor |
| chr2_+_234160217 | 0.03 |
ENST00000392017.4
ENST00000347464.5 ENST00000444735.1 ENST00000373525.5 ENST00000419681.1 |
ATG16L1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr7_-_22234381 | 0.03 |
ENST00000458533.1
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_-_207629997 | 0.03 |
ENST00000454776.2
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr17_+_76374714 | 0.02 |
ENST00000262764.6
ENST00000589689.1 ENST00000329897.7 ENST00000592043.1 ENST00000587356.1 |
PGS1
|
phosphatidylglycerophosphate synthase 1 |
| chr17_+_37356528 | 0.02 |
ENST00000225430.4
|
RPL19
|
ribosomal protein L19 |
| chr17_+_37356586 | 0.02 |
ENST00000579260.1
ENST00000582193.1 |
RPL19
|
ribosomal protein L19 |
| chr8_-_101965104 | 0.02 |
ENST00000437293.1
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_-_39968406 | 0.02 |
ENST00000393928.1
|
LEPREL4
|
leprecan-like 4 |
| chr2_+_119699864 | 0.02 |
ENST00000541757.1
ENST00000412481.1 |
MARCO
|
macrophage receptor with collagenous structure |
| chr9_-_131418944 | 0.02 |
ENST00000419989.1
ENST00000451652.1 ENST00000372715.2 |
WDR34
|
WD repeat domain 34 |
| chr5_-_133747589 | 0.02 |
ENST00000458198.2
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr10_-_43904608 | 0.02 |
ENST00000337970.3
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr17_+_37356555 | 0.02 |
ENST00000579374.1
|
RPL19
|
ribosomal protein L19 |
| chr19_-_40791302 | 0.02 |
ENST00000392038.2
ENST00000578123.1 |
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_+_39969183 | 0.01 |
ENST00000321562.4
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr11_-_126138808 | 0.01 |
ENST00000332118.6
ENST00000532259.1 |
SRPR
|
signal recognition particle receptor (docking protein) |
| chr16_+_67197288 | 0.01 |
ENST00000264009.8
ENST00000421453.1 |
HSF4
|
heat shock transcription factor 4 |
| chr3_+_127771212 | 0.01 |
ENST00000243253.3
ENST00000481210.1 |
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr11_+_6947720 | 0.01 |
ENST00000414517.2
|
ZNF215
|
zinc finger protein 215 |
| chr8_-_125384927 | 0.01 |
ENST00000297632.6
|
TMEM65
|
transmembrane protein 65 |
| chr17_-_39968855 | 0.01 |
ENST00000355468.3
ENST00000590496.1 |
LEPREL4
|
leprecan-like 4 |
| chr1_-_213031418 | 0.01 |
ENST00000356684.3
ENST00000426161.1 ENST00000424044.1 |
FLVCR1-AS1
|
FLVCR1 antisense RNA 1 (head to head) |
| chr11_+_6947647 | 0.01 |
ENST00000278319.5
|
ZNF215
|
zinc finger protein 215 |
| chr10_-_99393208 | 0.01 |
ENST00000307450.6
|
MORN4
|
MORN repeat containing 4 |
| chr14_+_64854958 | 0.01 |
ENST00000555709.2
ENST00000554739.1 ENST00000554768.1 ENST00000216605.8 |
MTHFD1
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr9_-_21142144 | 0.01 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr7_+_36429409 | 0.01 |
ENST00000265748.2
|
ANLN
|
anillin, actin binding protein |
| chr5_-_133747551 | 0.01 |
ENST00000395009.3
|
CDKN2AIPNL
|
CDKN2A interacting protein N-terminal like |
| chr6_-_170893669 | 0.01 |
ENST00000392090.2
ENST00000542896.1 ENST00000453163.2 ENST00000537445.1 |
PDCD2
|
programmed cell death 2 |
| chr4_-_163085141 | 0.01 |
ENST00000427802.2
ENST00000306100.5 |
FSTL5
|
follistatin-like 5 |
| chr4_+_75230853 | 0.01 |
ENST00000244869.2
|
EREG
|
epiregulin |
| chr11_+_61522844 | 0.01 |
ENST00000265460.5
|
MYRF
|
myelin regulatory factor |
| chr2_-_207630033 | 0.01 |
ENST00000449792.1
|
MDH1B
|
malate dehydrogenase 1B, NAD (soluble) |
| chr9_+_96793076 | 0.00 |
ENST00000375360.3
|
PTPDC1
|
protein tyrosine phosphatase domain containing 1 |
| chrX_-_132091284 | 0.00 |
ENST00000370833.2
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr6_+_74104471 | 0.00 |
ENST00000370336.4
|
DDX43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr1_+_26758790 | 0.00 |
ENST00000427245.2
ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS
|
dehydrodolichyl diphosphate synthase |
| chr6_+_3118926 | 0.00 |
ENST00000380379.5
|
BPHL
|
biphenyl hydrolase-like (serine hydrolase) |
| chr18_+_39766626 | 0.00 |
ENST00000593234.1
ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907
|
long intergenic non-protein coding RNA 907 |
| chr21_-_33104367 | 0.00 |
ENST00000286835.7
ENST00000399804.1 |
SCAF4
|
SR-related CTD-associated factor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX1_PAX9
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 0.2 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 0.0 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.0 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |