Project
A549 cells infected with SARS-CoV-2 Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for OTX1
Z-value: 0.55
Transcription factors associated with OTX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX1
|
ENSG00000115507.5 | orthodenticle homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OTX1 | hg19_v2_chr2_+_63277927_63277938 | 0.38 | 4.6e-01 | Click! |
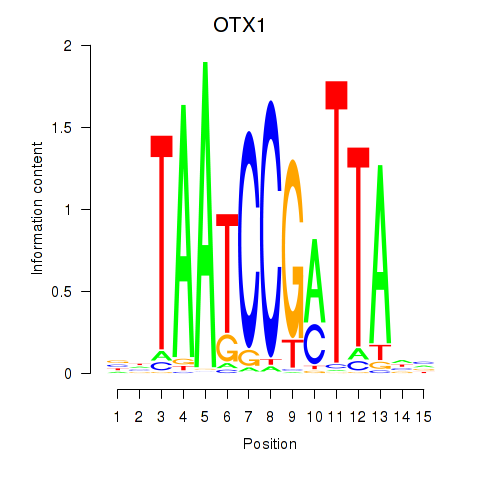
Activity profile of OTX1 motif
Sorted Z-values of OTX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_104211294 | 0.59 |
ENST00000239125.1
|
C10orf95
|
chromosome 10 open reading frame 95 |
| chr21_+_33671264 | 0.59 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr16_-_21452040 | 0.50 |
ENST00000521589.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr7_-_91509972 | 0.44 |
ENST00000425936.1
|
MTERF
|
mitochondrial transcription termination factor |
| chr2_-_14541060 | 0.28 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr19_-_46580352 | 0.22 |
ENST00000601672.1
|
IGFL4
|
IGF-like family member 4 |
| chr3_-_12587055 | 0.22 |
ENST00000564146.3
|
C3orf83
|
chromosome 3 open reading frame 83 |
| chr8_+_109455830 | 0.21 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr4_+_22999152 | 0.21 |
ENST00000511453.1
|
RP11-412P11.1
|
RP11-412P11.1 |
| chr14_-_65769392 | 0.20 |
ENST00000555736.1
|
CTD-2509G16.5
|
CTD-2509G16.5 |
| chr3_-_69062742 | 0.19 |
ENST00000424374.1
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr16_-_15950868 | 0.18 |
ENST00000396324.3
ENST00000452625.2 ENST00000576790.2 ENST00000300036.5 |
MYH11
|
myosin, heavy chain 11, smooth muscle |
| chr9_+_116172958 | 0.18 |
ENST00000374165.1
|
C9orf43
|
chromosome 9 open reading frame 43 |
| chr11_+_113258495 | 0.18 |
ENST00000303941.3
|
ANKK1
|
ankyrin repeat and kinase domain containing 1 |
| chr1_+_229440129 | 0.17 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr19_+_35940486 | 0.17 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr5_-_154230130 | 0.17 |
ENST00000519501.1
ENST00000518651.1 ENST00000517938.1 ENST00000520461.1 |
FAXDC2
|
fatty acid hydroxylase domain containing 2 |
| chr2_-_61389168 | 0.17 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr19_-_3761673 | 0.16 |
ENST00000316757.3
|
APBA3
|
amyloid beta (A4) precursor protein-binding, family A, member 3 |
| chr3_+_138340049 | 0.16 |
ENST00000464668.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr3_+_138340067 | 0.16 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr17_-_71223839 | 0.16 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr13_+_24825824 | 0.15 |
ENST00000434675.1
ENST00000494772.1 |
SPATA13
|
spermatogenesis associated 13 |
| chr3_+_142315294 | 0.15 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr9_-_35563896 | 0.15 |
ENST00000399742.2
|
FAM166B
|
family with sequence similarity 166, member B |
| chr5_+_134303591 | 0.15 |
ENST00000282611.6
|
CATSPER3
|
cation channel, sperm associated 3 |
| chr8_-_56986768 | 0.14 |
ENST00000523936.1
|
RPS20
|
ribosomal protein S20 |
| chr3_-_69062790 | 0.14 |
ENST00000540955.1
ENST00000456376.1 |
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr19_+_10531150 | 0.13 |
ENST00000352831.6
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr3_+_111393659 | 0.13 |
ENST00000477665.1
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr2_-_211090162 | 0.13 |
ENST00000233710.3
|
ACADL
|
acyl-CoA dehydrogenase, long chain |
| chr6_+_33359582 | 0.13 |
ENST00000450504.1
|
KIFC1
|
kinesin family member C1 |
| chr3_-_27410847 | 0.13 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr19_-_16008880 | 0.12 |
ENST00000011989.7
ENST00000221700.6 |
CYP4F2
|
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr1_+_28764653 | 0.12 |
ENST00000373836.3
|
PHACTR4
|
phosphatase and actin regulator 4 |
| chr16_+_14802801 | 0.12 |
ENST00000526520.1
ENST00000531598.2 |
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr19_-_42916499 | 0.12 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr5_-_54988448 | 0.11 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr9_-_95244781 | 0.11 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr3_+_188664988 | 0.11 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chrX_+_53123314 | 0.11 |
ENST00000605526.1
ENST00000604062.1 ENST00000604369.1 ENST00000366185.2 ENST00000604849.1 |
RP11-258C19.5
|
long intergenic non-protein coding RNA 1155 |
| chr19_-_47616992 | 0.11 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr12_-_107380846 | 0.10 |
ENST00000548101.1
ENST00000550496.1 ENST00000552029.1 |
MTERFD3
|
MTERF domain containing 3 |
| chr4_-_83769996 | 0.10 |
ENST00000511338.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr11_-_8959758 | 0.10 |
ENST00000531618.1
|
ASCL3
|
achaete-scute family bHLH transcription factor 3 |
| chr6_+_96025341 | 0.10 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr5_-_81574160 | 0.10 |
ENST00000510210.1
ENST00000512493.1 ENST00000507980.1 ENST00000511844.1 ENST00000510019.1 |
RPS23
|
ribosomal protein S23 |
| chr4_+_159727272 | 0.10 |
ENST00000379346.3
|
FNIP2
|
folliculin interacting protein 2 |
| chr9_+_123884038 | 0.10 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr19_-_41903161 | 0.10 |
ENST00000602129.1
ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5
|
exosome component 5 |
| chr1_-_247335269 | 0.10 |
ENST00000543802.2
ENST00000491356.1 ENST00000472531.1 ENST00000340684.6 |
ZNF124
|
zinc finger protein 124 |
| chr11_-_130786333 | 0.09 |
ENST00000533214.1
ENST00000528555.1 ENST00000530356.1 ENST00000539184.1 |
SNX19
|
sorting nexin 19 |
| chr8_-_133637624 | 0.09 |
ENST00000522789.1
|
LRRC6
|
leucine rich repeat containing 6 |
| chr4_+_159727222 | 0.09 |
ENST00000512986.1
|
FNIP2
|
folliculin interacting protein 2 |
| chr14_+_102196739 | 0.09 |
ENST00000556973.1
|
RP11-796G6.2
|
Uncharacterized protein |
| chr3_+_111393501 | 0.09 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr6_-_41039567 | 0.09 |
ENST00000468811.1
|
OARD1
|
O-acyl-ADP-ribose deacylase 1 |
| chr7_-_100895414 | 0.09 |
ENST00000435848.1
ENST00000474120.1 |
FIS1
|
fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) |
| chrX_-_48937684 | 0.08 |
ENST00000465382.1
ENST00000423215.2 |
WDR45
|
WD repeat domain 45 |
| chr22_-_28316116 | 0.08 |
ENST00000415296.1
|
PITPNB
|
phosphatidylinositol transfer protein, beta |
| chr2_+_204103733 | 0.08 |
ENST00000443941.1
|
CYP20A1
|
cytochrome P450, family 20, subfamily A, polypeptide 1 |
| chr2_+_232135245 | 0.08 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr5_-_54988559 | 0.08 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr19_-_36001386 | 0.08 |
ENST00000461300.1
|
DMKN
|
dermokine |
| chr22_+_39052632 | 0.08 |
ENST00000411557.1
ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1
|
chibby homolog 1 (Drosophila) |
| chr2_-_106054952 | 0.08 |
ENST00000336660.5
ENST00000393352.3 ENST00000607522.1 |
FHL2
|
four and a half LIM domains 2 |
| chr1_+_174844645 | 0.08 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr8_-_145652336 | 0.07 |
ENST00000529182.1
ENST00000526054.1 |
VPS28
|
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr19_-_55677920 | 0.07 |
ENST00000524407.2
ENST00000526003.1 ENST00000534170.1 |
DNAAF3
|
dynein, axonemal, assembly factor 3 |
| chr9_-_35665165 | 0.07 |
ENST00000343259.3
ENST00000378387.3 |
ARHGEF39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr11_-_65626753 | 0.07 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr5_-_66942617 | 0.07 |
ENST00000507298.1
|
RP11-83M16.5
|
RP11-83M16.5 |
| chr19_+_47616682 | 0.07 |
ENST00000594526.1
|
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr8_+_132916318 | 0.07 |
ENST00000254624.5
ENST00000522709.1 |
EFR3A
|
EFR3 homolog A (S. cerevisiae) |
| chr11_+_8704748 | 0.07 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr1_-_146696901 | 0.07 |
ENST00000369272.3
ENST00000441068.2 |
FMO5
|
flavin containing monooxygenase 5 |
| chr11_-_85376121 | 0.07 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr10_-_116418053 | 0.07 |
ENST00000277895.5
|
ABLIM1
|
actin binding LIM protein 1 |
| chr6_+_42018614 | 0.07 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr15_+_58702742 | 0.07 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr7_+_99195677 | 0.07 |
ENST00000431679.1
|
GS1-259H13.2
|
GS1-259H13.2 |
| chr22_-_39052300 | 0.06 |
ENST00000355830.6
|
FAM227A
|
family with sequence similarity 227, member A |
| chr2_-_96192450 | 0.06 |
ENST00000609975.1
|
RP11-440D17.3
|
RP11-440D17.3 |
| chr9_+_67968793 | 0.06 |
ENST00000417488.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr5_+_59783540 | 0.06 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr14_-_27066636 | 0.06 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr6_-_144416737 | 0.06 |
ENST00000367569.2
|
SF3B5
|
splicing factor 3b, subunit 5, 10kDa |
| chr10_-_28571015 | 0.06 |
ENST00000375719.3
ENST00000375732.1 |
MPP7
|
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr15_-_43877062 | 0.06 |
ENST00000381885.1
ENST00000396923.3 |
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr11_-_65626797 | 0.06 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr10_-_71993138 | 0.06 |
ENST00000608321.1
|
PPA1
|
pyrophosphatase (inorganic) 1 |
| chr5_-_39425290 | 0.06 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr3_+_142315225 | 0.06 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr2_+_110656005 | 0.06 |
ENST00000437679.2
|
LIMS3
|
LIM and senescent cell antigen-like domains 3 |
| chr12_-_14956396 | 0.05 |
ENST00000535328.1
ENST00000261167.2 |
WBP11
|
WW domain binding protein 11 |
| chr6_+_147981838 | 0.05 |
ENST00000427015.1
ENST00000432506.1 |
RP11-307P5.1
|
RP11-307P5.1 |
| chr11_-_67210930 | 0.05 |
ENST00000453768.2
ENST00000545016.1 ENST00000341356.5 |
CORO1B
|
coronin, actin binding protein, 1B |
| chr11_-_124767693 | 0.05 |
ENST00000533054.1
|
ROBO4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr5_-_39425222 | 0.05 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr5_+_101569696 | 0.05 |
ENST00000597120.1
|
AC008948.1
|
AC008948.1 |
| chr3_-_156878540 | 0.05 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr1_+_150293921 | 0.05 |
ENST00000324862.6
|
PRPF3
|
pre-mRNA processing factor 3 |
| chr4_+_76439095 | 0.05 |
ENST00000506261.1
|
THAP6
|
THAP domain containing 6 |
| chr1_+_243419306 | 0.05 |
ENST00000355875.4
ENST00000391846.1 ENST00000366541.3 ENST00000343783.6 |
SDCCAG8
|
serologically defined colon cancer antigen 8 |
| chr2_-_85637459 | 0.05 |
ENST00000409921.1
|
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr9_+_128509624 | 0.05 |
ENST00000342287.5
ENST00000373487.4 |
PBX3
|
pre-B-cell leukemia homeobox 3 |
| chr1_+_178310581 | 0.05 |
ENST00000462775.1
|
RASAL2
|
RAS protein activator like 2 |
| chr8_+_109455845 | 0.05 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr14_-_75083313 | 0.04 |
ENST00000556652.1
ENST00000555313.1 |
CTD-2207P18.2
|
CTD-2207P18.2 |
| chr1_+_241695670 | 0.04 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr5_+_80256453 | 0.04 |
ENST00000265080.4
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2 |
| chr2_-_201936302 | 0.04 |
ENST00000453765.1
ENST00000452799.1 ENST00000446678.1 ENST00000418596.3 |
FAM126B
|
family with sequence similarity 126, member B |
| chr1_+_160709076 | 0.04 |
ENST00000359331.4
ENST00000495334.1 |
SLAMF7
|
SLAM family member 7 |
| chr8_-_110988070 | 0.04 |
ENST00000524391.1
|
KCNV1
|
potassium channel, subfamily V, member 1 |
| chrX_-_138072729 | 0.04 |
ENST00000455663.1
|
FGF13
|
fibroblast growth factor 13 |
| chr1_-_161039753 | 0.04 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr11_-_111957451 | 0.04 |
ENST00000504148.2
ENST00000541231.1 |
TIMM8B
|
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr1_-_160924589 | 0.04 |
ENST00000368029.3
|
ITLN2
|
intelectin 2 |
| chr15_+_42787452 | 0.04 |
ENST00000249647.3
|
SNAP23
|
synaptosomal-associated protein, 23kDa |
| chr20_+_3052264 | 0.04 |
ENST00000217386.2
|
OXT
|
oxytocin/neurophysin I prepropeptide |
| chrX_+_100645812 | 0.04 |
ENST00000427805.2
ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A
RPL36A-HNRNPH2
|
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr11_-_78052923 | 0.04 |
ENST00000340149.2
|
GAB2
|
GRB2-associated binding protein 2 |
| chr9_-_95298314 | 0.04 |
ENST00000344604.5
ENST00000375540.1 |
ECM2
|
extracellular matrix protein 2, female organ and adipocyte specific |
| chr8_+_143781513 | 0.04 |
ENST00000292430.6
ENST00000561179.1 ENST00000518841.1 ENST00000519387.1 |
LY6K
|
lymphocyte antigen 6 complex, locus K |
| chr14_-_102706934 | 0.04 |
ENST00000523231.1
ENST00000524370.1 ENST00000517966.1 |
MOK
|
MOK protein kinase |
| chr3_+_42544084 | 0.04 |
ENST00000543411.1
ENST00000438259.2 ENST00000439731.1 ENST00000325123.4 |
VIPR1
|
vasoactive intestinal peptide receptor 1 |
| chr17_-_36981556 | 0.04 |
ENST00000536127.1
ENST00000225428.5 |
CWC25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
| chr17_-_79623597 | 0.04 |
ENST00000574024.1
ENST00000331056.5 |
PDE6G
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr7_+_1609765 | 0.03 |
ENST00000437964.1
ENST00000533935.1 ENST00000532358.1 ENST00000524978.1 |
PSMG3-AS1
|
PSMG3 antisense RNA 1 (head to head) |
| chr10_+_114133773 | 0.03 |
ENST00000354655.4
|
ACSL5
|
acyl-CoA synthetase long-chain family member 5 |
| chr4_+_100737954 | 0.03 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr10_+_24755416 | 0.03 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr5_+_147443534 | 0.03 |
ENST00000398454.1
ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5
|
serine peptidase inhibitor, Kazal type 5 |
| chr16_-_57880439 | 0.03 |
ENST00000565684.1
|
KIFC3
|
kinesin family member C3 |
| chr1_+_109756523 | 0.03 |
ENST00000234677.2
ENST00000369923.4 |
SARS
|
seryl-tRNA synthetase |
| chr6_-_34393825 | 0.03 |
ENST00000605528.1
ENST00000326199.8 |
RPS10-NUDT3
RPS10
|
RPS10-NUDT3 readthrough ribosomal protein S10 |
| chr7_+_55980331 | 0.03 |
ENST00000429591.2
|
ZNF713
|
zinc finger protein 713 |
| chr21_-_43816052 | 0.03 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr14_-_70040339 | 0.03 |
ENST00000599174.1
|
CCDC177
|
Homo sapiens coiled-coil domain containing 177 (CCDC177), mRNA. |
| chr2_-_112642267 | 0.03 |
ENST00000341068.3
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr11_-_67275542 | 0.03 |
ENST00000531506.1
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr1_+_184020830 | 0.03 |
ENST00000533373.1
ENST00000423085.2 |
TSEN15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr2_-_190448118 | 0.03 |
ENST00000440626.1
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr15_+_75550940 | 0.03 |
ENST00000300576.5
|
GOLGA6C
|
golgin A6 family, member C |
| chr12_-_71533055 | 0.03 |
ENST00000552128.1
|
TSPAN8
|
tetraspanin 8 |
| chr12_+_57881740 | 0.03 |
ENST00000262027.5
ENST00000315473.5 |
MARS
|
methionyl-tRNA synthetase |
| chr3_-_69062764 | 0.03 |
ENST00000295571.5
|
EOGT
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr1_+_40840320 | 0.03 |
ENST00000372708.1
|
SMAP2
|
small ArfGAP2 |
| chr2_+_102721023 | 0.03 |
ENST00000409589.1
ENST00000409329.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr1_+_87458692 | 0.02 |
ENST00000370548.2
ENST00000356813.4 |
RP5-1052I5.2
HS2ST1
|
Heparan sulfate 2-O-sulfotransferase 1 heparan sulfate 2-O-sulfotransferase 1 |
| chr1_+_241695424 | 0.02 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_-_74619152 | 0.02 |
ENST00000440727.1
ENST00000409240.1 |
DCTN1
|
dynactin 1 |
| chr19_-_36001113 | 0.02 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr15_-_61521495 | 0.02 |
ENST00000335670.6
|
RORA
|
RAR-related orphan receptor A |
| chr6_+_150070857 | 0.02 |
ENST00000544496.1
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr16_-_21436459 | 0.02 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr18_-_70532906 | 0.02 |
ENST00000299430.2
ENST00000397929.1 |
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr7_+_128379449 | 0.02 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr3_+_11314072 | 0.02 |
ENST00000444619.1
|
ATG7
|
autophagy related 7 |
| chr1_-_78444738 | 0.02 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr1_+_221054411 | 0.02 |
ENST00000427693.1
|
HLX
|
H2.0-like homeobox |
| chr2_+_234580499 | 0.02 |
ENST00000354728.4
|
UGT1A9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr5_-_59783882 | 0.02 |
ENST00000505507.2
ENST00000502484.2 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr5_+_96038476 | 0.02 |
ENST00000511049.1
ENST00000309190.5 ENST00000510156.1 ENST00000509903.1 ENST00000511782.1 ENST00000504465.1 |
CAST
|
calpastatin |
| chr3_-_141747950 | 0.02 |
ENST00000497579.1
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_-_52002403 | 0.02 |
ENST00000490063.1
ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4
|
poly(rC) binding protein 4 |
| chr9_-_100684845 | 0.02 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr5_+_140079919 | 0.02 |
ENST00000274712.3
|
ZMAT2
|
zinc finger, matrin-type 2 |
| chr12_+_6603253 | 0.02 |
ENST00000382457.4
ENST00000545962.1 |
NCAPD2
|
non-SMC condensin I complex, subunit D2 |
| chrX_+_149861836 | 0.02 |
ENST00000542156.1
ENST00000370390.3 ENST00000490316.2 ENST00000445323.2 ENST00000544228.1 ENST00000451863.2 |
MTMR1
|
myotubularin related protein 1 |
| chr16_+_33204156 | 0.02 |
ENST00000398667.4
|
TP53TG3C
|
TP53 target 3C |
| chr3_-_138763734 | 0.02 |
ENST00000413199.1
ENST00000502927.2 |
PRR23C
|
proline rich 23C |
| chr19_+_41903709 | 0.02 |
ENST00000542943.1
ENST00000457836.2 |
BCKDHA
|
branched chain keto acid dehydrogenase E1, alpha polypeptide |
| chr16_-_69448 | 0.02 |
ENST00000326592.9
|
WASH4P
|
WAS protein family homolog 4 pseudogene |
| chr6_+_26251835 | 0.02 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr1_-_36930012 | 0.02 |
ENST00000373116.5
|
MRPS15
|
mitochondrial ribosomal protein S15 |
| chr10_-_35104185 | 0.02 |
ENST00000374789.3
ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3
|
par-3 family cell polarity regulator |
| chr11_+_128563652 | 0.02 |
ENST00000527786.2
|
FLI1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_-_89653576 | 0.01 |
ENST00000420869.1
|
TRIM49D1
|
tripartite motif containing 49D1 |
| chr20_-_33539655 | 0.01 |
ENST00000451957.2
|
GSS
|
glutathione synthetase |
| chr7_+_120702819 | 0.01 |
ENST00000423795.1
|
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr18_+_8717369 | 0.01 |
ENST00000359865.3
ENST00000400050.3 ENST00000306285.7 |
SOGA2
|
SOGA family member 2 |
| chr1_-_151300160 | 0.01 |
ENST00000368874.4
|
PI4KB
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr5_+_135496675 | 0.01 |
ENST00000507637.1
|
SMAD5
|
SMAD family member 5 |
| chrX_-_148571884 | 0.01 |
ENST00000537071.1
|
IDS
|
iduronate 2-sulfatase |
| chr22_-_36903101 | 0.01 |
ENST00000397224.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr2_+_122513109 | 0.01 |
ENST00000389682.3
ENST00000536142.1 |
TSN
|
translin |
| chr11_-_67276100 | 0.01 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr2_+_234580525 | 0.01 |
ENST00000609637.1
|
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr3_+_11314099 | 0.01 |
ENST00000446450.2
ENST00000354956.5 ENST00000354449.3 ENST00000419112.1 |
ATG7
|
autophagy related 7 |
| chr6_-_131211534 | 0.01 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr3_-_193272874 | 0.01 |
ENST00000342695.4
|
ATP13A4
|
ATPase type 13A4 |
| chr5_+_96038554 | 0.01 |
ENST00000508197.1
|
CAST
|
calpastatin |
| chr2_-_74618964 | 0.01 |
ENST00000417090.1
ENST00000409868.1 |
DCTN1
|
dynactin 1 |
| chr18_-_59274139 | 0.01 |
ENST00000586949.1
|
RP11-879F14.2
|
RP11-879F14.2 |
| chr16_-_21868978 | 0.01 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr1_+_17531614 | 0.01 |
ENST00000375471.4
|
PADI1
|
peptidyl arginine deiminase, type I |
| chr1_-_186430222 | 0.01 |
ENST00000391997.2
|
PDC
|
phosducin |
| chr1_-_160832490 | 0.01 |
ENST00000322302.7
ENST00000368033.3 |
CD244
|
CD244 molecule, natural killer cell receptor 2B4 |
| chr16_-_58163299 | 0.01 |
ENST00000262498.3
|
C16orf80
|
chromosome 16 open reading frame 80 |
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.2 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:2000395 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0071964 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:1904578 | response to thapsigargin(GO:1904578) cellular response to thapsigargin(GO:1904579) |
| 0.0 | 0.0 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.0 | GO:0045925 | positive regulation of female receptivity(GO:0045925) positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.0 | GO:0039521 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.0 | 0.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031780 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.1 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |